5NCT
 
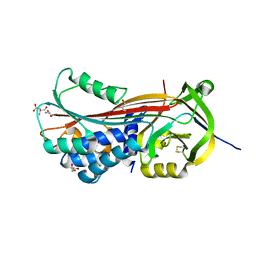 | | Structure of the trypsin induced serpin-type proteinase inhibitor, miropin. | | Descriptor: | ASPARTIC ACID, GLYCEROL, SERINE, ... | | Authors: | Goulas, T, Ksiazek, M, Garcia-Ferrer, I, Mizgalska, D, Potempa, J, Gomis-Ruth, X. | | Deposit date: | 2017-03-06 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A structure-derived snap-trap mechanism of a multispecific serpin from the dysbiotic human oral microbiome.
J. Biol. Chem., 292, 2017
|
|
5NCU
 
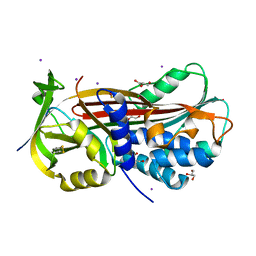 | | Structure of the subtilisin induced serpin-type proteinase inhibitor, miropin. | | Descriptor: | CHLORIDE ION, GLYCEROL, IODIDE ION, ... | | Authors: | Goulas, T, Ksiazek, M, Garcia-Ferrer, I, Mizgalska, D, Potempa, J, Gomis-Ruth, X. | | Deposit date: | 2017-03-06 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A structure-derived snap-trap mechanism of a multispecific serpin from the dysbiotic human oral microbiome.
J. Biol. Chem., 292, 2017
|
|
5NCW
 
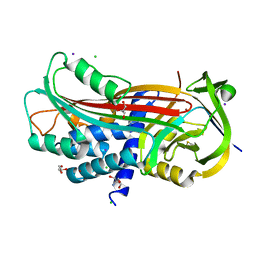 | | Structure of the trypsin induced serpin-type proteinase inhibitor, miropin (V367K/K368A mutant). | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Goulas, T, Ksiazek, M, Garcia-Ferrer, I, Mizgalska, D, Potempa, J, Gomis-Ruth, X. | | Deposit date: | 2017-03-06 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A structure-derived snap-trap mechanism of a multispecific serpin from the dysbiotic human oral microbiome.
J. Biol. Chem., 292, 2017
|
|
5NCS
 
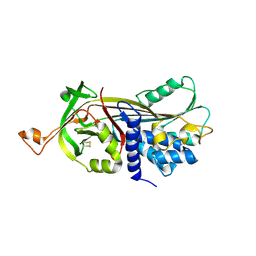 | | Structure of the native serpin-type proteinase inhibitor, miropin. | | Descriptor: | Serpin | | Authors: | Goulas, T, Ksiazek, M, Garcia-Ferrer, I, Mizgalska, D, Potempa, J, Gomis-Ruth, X. | | Deposit date: | 2017-03-06 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A structure-derived snap-trap mechanism of a multispecific serpin from the dysbiotic human oral microbiome.
J. Biol. Chem., 292, 2017
|
|
1OKR
 
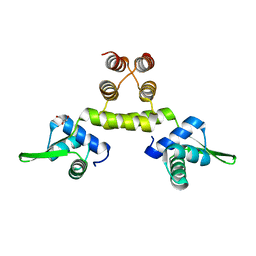 | | Three-dimensional structure of S.aureus methicillin-resistance regulating transcriptional repressor MecI. | | Descriptor: | CHLORIDE ION, GLYCEROL, METHICILLIN RESISTANCE REGULATORY PROTEIN MECI | | Authors: | Garcia-Castellanos, R, Marrero, A, Mallorqui-Fernandez, G, Potempa, J, Coll, M, Gomis-Ruth, F.X. | | Deposit date: | 2003-07-28 | | Release date: | 2003-10-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Three-Dimensional Structure of Meci: Molecular Basis for Transcriptional Regulation of Staphylococcal Methicillin Resistance
J.Biol.Chem., 278, 2003
|
|
1AU8
 
 | | HUMAN CATHEPSIN G | | Descriptor: | CATHEPSIN G, N-(3-carboxypropanoyl)-L-valyl-N-[(1R)-5-amino-1-phosphonopentyl]-L-prolinamide | | Authors: | Medrano, F.J, Bode, W, Banbula, A, Potempa, J. | | Deposit date: | 1997-09-12 | | Release date: | 1998-10-14 | | Last modified: | 2012-12-12 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | HUMAN CATHEPSIN G
to be published
|
|
6I9A
 
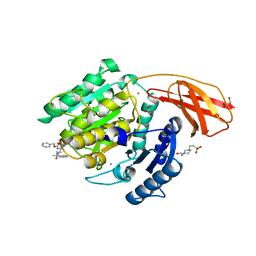 | | Porphyromonas gingivalis gingipain K (Kgp) in complex with inhibitor KYT-36 | | Descriptor: | (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Gomis-Ruth, F.X, Guevara, T, Rofdriguez-Banqueri, A. | | Deposit date: | 2018-11-22 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural determinants of inhibition of Porphyromonas gingivalis gingipain K by KYT-36, a potent, selective, and bioavailable peptidase inhibitor.
Sci Rep, 9, 2019
|
|
7NT4
 
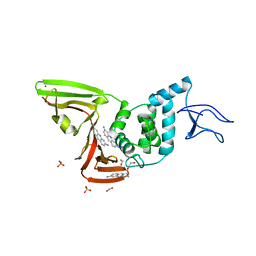 | | X-ray structure of SCoV2-PLpro in complex with small molecule inhibitor | | Descriptor: | 1,2-ETHANEDIOL, Non-structural protein 3, PROFLAVIN, ... | | Authors: | Napolitano, V, Mourao, A, Bostock, M, Matsuda, A, Czarna, A, Popowicz, G.M. | | Deposit date: | 2021-03-09 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Acriflavine, a clinically approved drug, inhibits SARS-CoV-2 and other betacoronaviruses.
Cell Chem Biol, 29, 2022
|
|
7OD0
 
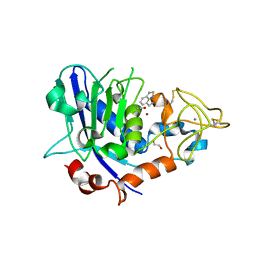 | | Mirolysin in complex with compound 9 | | Descriptor: | 1,2-ETHANEDIOL, 2,1,3-benzothiadiazol-4-ylmethanamine, ACETATE ION, ... | | Authors: | Zak, K.M, Bostock, M.J, Ksiazek, M. | | Deposit date: | 2021-04-28 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Latency, thermal stability, and identification of an inhibitory compound of mirolysin, a secretory protease of the human periodontopathogen Tannerella forsythia .
J Enzyme Inhib Med Chem, 36, 2021
|
|
6YZG
 
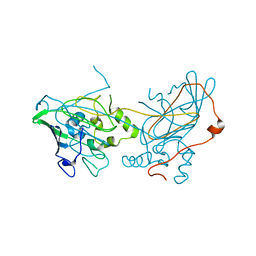 | |
4ZFI
 
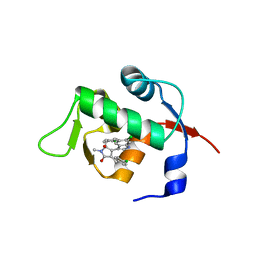 | | Structure of Mdm2 with low molecular weight inhibitor | | Descriptor: | (5S)-3,5-bis(4-chlorobenzyl)-4-(6-chloro-1H-indol-3-yl)-5-hydroxy-1-methyl-1,5-dihydro-2H-pyrrol-2-one, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Zak, K.M, Twarda-Clapa, A, Wrona, E.M, Grudnik, P, Dubin, G, Holak, T.A. | | Deposit date: | 2015-04-21 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Unique Mdm2-Binding Mode of the 3-Pyrrolin-2-one- and 2-Furanone-Based Antagonists of the p53-Mdm2 Interaction.
ACS Chem. Biol., 11, 2016
|
|
4ZGK
 
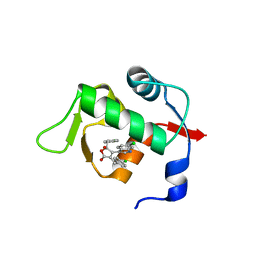 | | Structure of Mdm2 with low molecular weight inhibitor. | | Descriptor: | (5R)-3,5-bis(4-chlorobenzyl)-4-(6-chloro-1H-indol-3-yl)-5-hydroxyfuran-2(5H)-one, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Twarda-Clapa, A, Zak, K.M, Wrona, E.M, Grudnik, P, Dubin, G, Holak, T.A. | | Deposit date: | 2015-04-23 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Unique Mdm2-Binding Mode of the 3-Pyrrolin-2-one- and 2-Furanone-Based Antagonists of the p53-Mdm2 Interaction.
ACS Chem. Biol., 11, 2016
|
|
1CGH
 
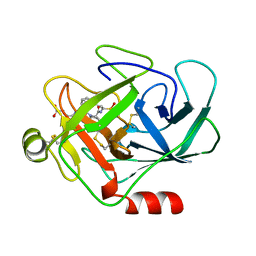 | | Human cathepsin G | | Descriptor: | CATHEPSIN G, N-(3-carboxypropanoyl)-L-valyl-N-{(1R)-1-[(S)-hydroxy(oxido)phosphanyl]-2-phenylethyl}-L-prolinamide | | Authors: | Hof, P, Bode, W. | | Deposit date: | 1996-06-26 | | Release date: | 1997-07-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The 1.8 A crystal structure of human cathepsin G in complex with Suc-Val-Pro-PheP-(OPh)2: a Janus-faced proteinase with two opposite specificities.
EMBO J., 15, 1996
|
|
1CVR
 
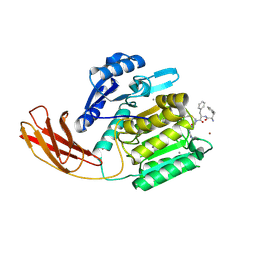 | |
8EHB
 
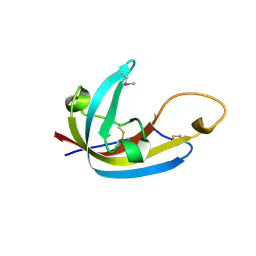 | |
8EHC
 
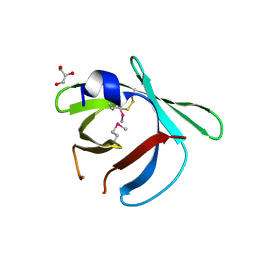 | |
8EHD
 
 | | Structure of Tannerella forsythia potempin E | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Potempin E (PotE) | | Authors: | Gomis-Ruth, F.X. | | Deposit date: | 2022-09-14 | | Release date: | 2022-12-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A unique network of attack, defence and competence on the outer membrane of the periodontitis pathogen Tannerella forsythia.
Chem Sci, 14, 2023
|
|
8EHE
 
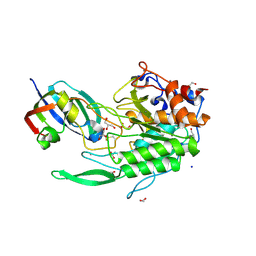 | |
5O0I
 
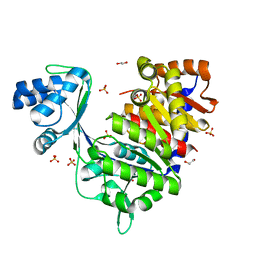 | | ADP-dependent glucokinase from Pyrococcus horikoshii | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, ADP-dependent glucokinase, ... | | Authors: | Grudnik, P, Dubin, G. | | Deposit date: | 2017-05-16 | | Release date: | 2018-05-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for ADP-dependent glucokinase inhibition by 8-bromo-substituted adenosine nucleotide.
J. Biol. Chem., 293, 2018
|
|
5O0J
 
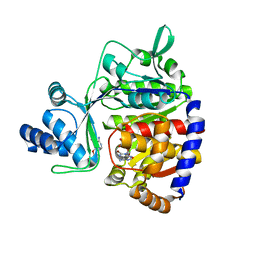 | | ADP-dependent glucokinase from Pyrococcus horikoshii | | Descriptor: | 8-BROMO-ADENOSINE-5'-MONOPHOSPHATE, ADP-dependent glucokinase, alpha-D-glucopyranose | | Authors: | Grudnik, P, Dubin, G. | | Deposit date: | 2017-05-16 | | Release date: | 2018-05-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural basis for ADP-dependent glucokinase inhibition by 8-bromo-substituted adenosine nucleotide.
J. Biol. Chem., 293, 2018
|
|
1T32
 
 | | A Dual Inhibitor of the Leukocyte Proteases Cathepsin G and Chymase with Therapeutic Efficacy in Animals Models of Inflammation | | Descriptor: | 2-[3-({METHYL[1-(2-NAPHTHOYL)PIPERIDIN-4-YL]AMINO}CARBONYL)-2-NAPHTHYL]-1-(1-NAPHTHYL)-2-OXOETHYLPHOSPHONIC ACID, Cathepsin G, SULFATE ION | | Authors: | de Garavilla, L, Greco, M.N, Giardino, E.C, Wells, G.I, Haertlein, B.J, Kauffman, J.A, Corcoran, T.W, Derian, C.K, Eckardt, A.J, Abraham, W.M, Sukumar, N, Chen, Z, Pineda, A.O, Mathews, F.S, Di Cera, E, Andrade-Gordon, P, Damiano, B.P, Maryanoff, B.E. | | Deposit date: | 2004-04-23 | | Release date: | 2005-03-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A novel, potent dual inhibitor of the leukocyte proteases cathepsin G and chymase: molecular mechanisms and anti-inflammatory activity in vivo.
J.Biol.Chem., 280, 2005
|
|
6SML
 
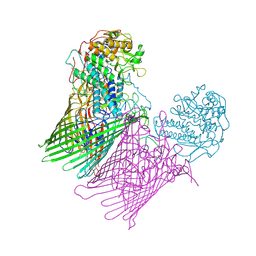 | | Structure of the RagAB peptide importer in the 'open-open' state | | Descriptor: | (1R,4S,6R)-6-({[2-(ACETYLAMINO)-2-DEOXY-ALPHA-D-GLUCOPYRANOSYL]OXY}METHYL)-4-HYDROXY-1-{[(15-METHYLHEXADECANOYL)OXY]METHYL}-4-OXIDO-7-OXO-3,5-DIOXA-8-AZA-4-PHOSPHAHEPTACOS-1-YL 15-METHYLHEXADECANOATE, GLY-THR-GLY-GLY-SER-THR-GLY-THR-THR-SER-ALA-GLY, Lipoprotein RagB, ... | | Authors: | White, J.B.R, Ranson, N.A, van den Berg, B. | | Deposit date: | 2019-08-22 | | Release date: | 2020-05-20 | | Last modified: | 2020-08-05 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural and functional insights into oligopeptide acquisition by the RagAB transporter from Porphyromonas gingivalis.
Nat Microbiol, 5, 2020
|
|
6SLJ
 
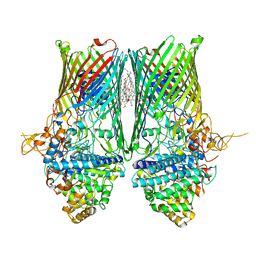 | | Structure of the RagAB peptide transporter | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, ALA-SER-THR-THR-GLY-ALA-ASN-SER-GLN-ARG, ... | | Authors: | Madej, M, Ranson, N.A, White, J.B.R. | | Deposit date: | 2019-08-20 | | Release date: | 2020-05-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Structural and functional insights into oligopeptide acquisition by the RagAB transporter from Porphyromonas gingivalis.
Nat Microbiol, 5, 2020
|
|
6SLI
 
 | | Structure of the RagAB peptide transporter | | Descriptor: | (1R,4S,6R)-6-({[2-(ACETYLAMINO)-2-DEOXY-ALPHA-D-GLUCOPYRANOSYL]OXY}METHYL)-4-HYDROXY-1-{[(15-METHYLHEXADECANOYL)OXY]METHYL}-4-OXIDO-7-OXO-3,5-DIOXA-8-AZA-4-PHOSPHAHEPTACOS-1-YL 15-METHYLHEXADECANOATE, (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, ALA-SER-THR-THR-GLY-GLY-ASN-SER-GLN-ARG-GLY-SER-GLY, ... | | Authors: | Madej, M, Ranson, N.A, White, J.B.R. | | Deposit date: | 2019-08-19 | | Release date: | 2020-05-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.38 Å) | | Cite: | Structural and functional insights into oligopeptide acquisition by the RagAB transporter from Porphyromonas gingivalis.
Nat Microbiol, 5, 2020
|
|
6SLN
 
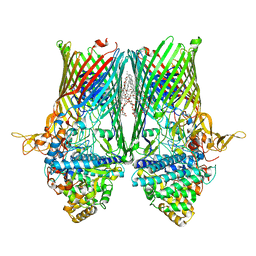 | | Structure of the RagAB peptide transporter | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Madej, M, Ranson, N.A, White, J.B.R. | | Deposit date: | 2019-08-20 | | Release date: | 2020-05-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural and functional insights into oligopeptide acquisition by the RagAB transporter from Porphyromonas gingivalis.
Nat Microbiol, 5, 2020
|
|
