2UZA
 
 | | CRYSTAL STRUCTURE OF THE FREE RADICAL INTERMEDIATE OF PYRUVATE:FERREDOXIN OXIDOREDUCTASE FROM DESULFOVIBRIO AFRICANUS | | Descriptor: | 2-ACETYL-THIAMINE DIPHOSPHATE, CALCIUM ION, CARBON DIOXIDE, ... | | Authors: | Chabriere, E, Cavazza, C, Contreras-Martel, C, Fontecilla-Camps, J.C. | | Deposit date: | 2007-04-27 | | Release date: | 2008-05-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Flexibility of thiamine diphosphate revealed by kinetic crystallographic studies of the reaction of pyruvate-ferredoxin oxidoreductase with pyruvate.
Structure, 14, 2006
|
|
2V3Q
 
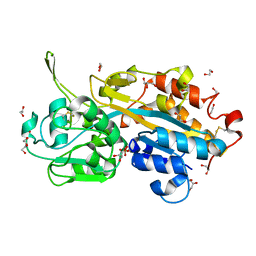 | | Serendipitous discovery and X-ray structure of a human phosphate binding apolipoprotein | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, HUMAN PHOSPHATE BINDING PROTEIN, ... | | Authors: | Morales, R, Berna, A, Carpentier, P, Elias, M, Contreras-Martel, C, Renault, F, Nicodeme, M, Chesne-Seck, M.-L, Bernier, F, Dupuy, J, Schaeffer, C, Diemer, H, Van Dorsselaer, A, Fontecilla, J.C, Masson, P, Rochu, D, Chabriere, E. | | Deposit date: | 2007-06-20 | | Release date: | 2008-07-22 | | Last modified: | 2016-01-27 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Tandem Use of X-Ray Crystallography and Mass Spectrometry to Obtain Ab Initio the Complete and Exact Amino Acids Sequence of Hpbp, a Human 38kDa Apolipoprotein
Proteins: Struct., Funct., Bioinf., 71, 2008
|
|
2C6W
 
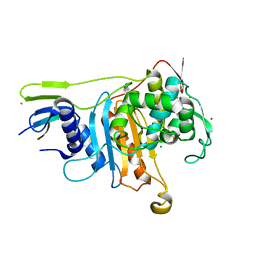 | | PENICILLIN-BINDING PROTEIN 1A (PBP-1A) FROM STREPTOCOCCUS PNEUMONIAE | | Descriptor: | CHLORIDE ION, PENICILLIN-BINDING PROTEIN 1A, ZINC ION | | Authors: | Contreras-Martel, C, Job, V, Di Guilmi, A.-M, Vernet, T, Dideberg, O, Dessen, A. | | Deposit date: | 2005-11-14 | | Release date: | 2005-12-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Crystal Structure of Penicillin-Binding Protein 1A (Pbp1A) Reveals a Mutational Hotspot Implicated in Beta-Lactam Resistance in Streptococcus Pneumoniae.
J.Mol.Biol., 355, 2006
|
|
2C5W
 
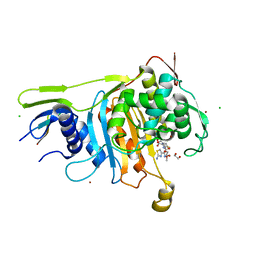 | | PENICILLIN-BINDING PROTEIN 1A (PBP-1A) ACYL-ENZYME COMPLEX (CEFOTAXIME) FROM STREPTOCOCCUS PNEUMONIAE | | Descriptor: | 1,2-ETHANEDIOL, CEFOTAXIME, C3' cleaved, ... | | Authors: | Contreras-Martel, C, Job, V, Di Guilmi, A.-M, Vernet, T, Dideberg, O, Dessen, A. | | Deposit date: | 2005-11-02 | | Release date: | 2005-12-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal Structure of Penicillin-Binding Protein 1A (Pbp1A) Reveals a Mutational Hotspot Implicated in Beta-Lactam Resistance in Streptococcus Pneumoniae.
J.Mol.Biol., 355, 2006
|
|
2WAE
 
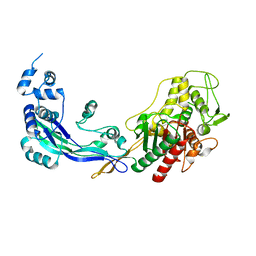 | | PENICILLIN-BINDING PROTEIN 2B (PBP-2B) FROM STREPTOCOCCUS PNEUMONIAE (STRAIN 5204) | | Descriptor: | PENICILLIN-BINDING PROTEIN 2B, ZINC ION | | Authors: | Contreras-Martel, C, Dahout-Gonzalez, C, Dos-Santos-Martins, A, Kotnik, M, Dessen, A. | | Deposit date: | 2009-02-05 | | Release date: | 2009-02-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Pbp Active Site Flexibility as the Key Mechanism for Beta-Lactam Resistance in Pneumococci
J.Mol.Biol., 387, 2009
|
|
2WAD
 
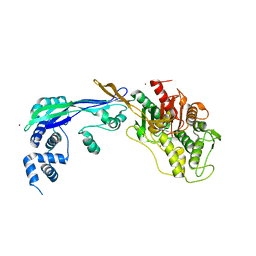 | | PENICILLIN-BINDING PROTEIN 2B (PBP-2B) FROM STREPTOCOCCUS PNEUMONIAE (STRAIN 5204) | | Descriptor: | PENICILLIN-BINDING PROTEIN 2B, ZINC ION | | Authors: | Contreras-Martel, C, Dahout-Gonzalez, C, Dos-Santos-Martins, A, Kotnik, M, Dessen, A. | | Deposit date: | 2009-02-05 | | Release date: | 2009-02-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Pbp Active Site Flexibility as the Key Mechanism for Beta-Lactam Resistance in Pneumococci
J.Mol.Biol., 387, 2009
|
|
2WAF
 
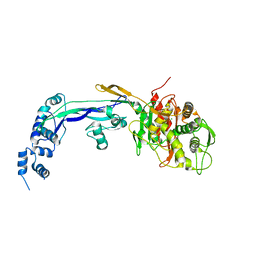 | | PENICILLIN-BINDING PROTEIN 2B (PBP-2B) FROM STREPTOCOCCUS PNEUMONIAE (STRAIN R6) | | Descriptor: | CHLORIDE ION, PENICILLIN-BINDING PROTEIN 2B | | Authors: | Contreras-Martel, C, Dahout-Gonzalez, C, Dos-Santos-Martins, A, Kotnik, M, Dessen, A. | | Deposit date: | 2009-02-05 | | Release date: | 2009-02-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Pbp Active Site Flexibility as the Key Mechanism for Beta-Lactam Resistance in Pneumococci
J.Mol.Biol., 387, 2009
|
|
2W1J
 
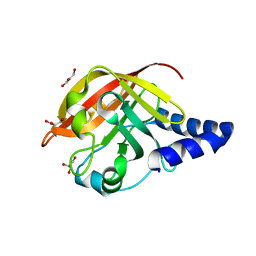 | | CRYSTAL STRUCTURE OF SORTASE C-1 (SRTC-1) from Streptococcus pneumoniae | | Descriptor: | GLYCEROL, PUTATIVE SORTASE | | Authors: | Manzano, C, Contreras-Martel, C, El Mortaji, L, Izore, T, Fenel, D, Vernet, T, Schoehn, G, Di Guilmi, A.M, Dessen, A. | | Deposit date: | 2008-10-17 | | Release date: | 2008-12-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Sortase-Mediated Pilus Fiber Biogenesis in Streptococcus Pneumoniae.
Structure, 16, 2008
|
|
2W1K
 
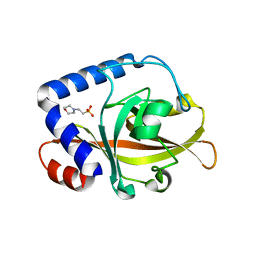 | | Crystal Structure of Sortase C-3 (SRTC-3) from Streptococcus pneumoniae | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, PUTATIVE SORTASE | | Authors: | Manzano, C, Contreras-Martel, C, El Mortaji, L, Izore, T, Fenel, D, Vernet, T, Schoehn, G, Di Guilmi, A.M, Dessen, A. | | Deposit date: | 2008-10-17 | | Release date: | 2008-12-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Sortase-Mediated Pilus Fiber Biogenesis in Streptococcus Pneumoniae.
Structure, 16, 2008
|
|
2WW8
 
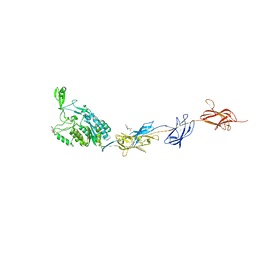 | | Structure of the pilus adhesin (RrgA) from Streptococcus pneumoniae | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, CELL WALL SURFACE ANCHOR FAMILY PROTEIN, ... | | Authors: | Izore, T, Contreras-Martel, C, El-Mortaji, L, Manzano, C, Terrasse, R, Vernet, T, Di-Guilmi, A.M, Dessen, A. | | Deposit date: | 2009-10-22 | | Release date: | 2010-01-19 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis of Host Cell Recognition by the Pilus Adhesin from Streptococcus Pneumoniae
Structure, 18, 2010
|
|
2Y1V
 
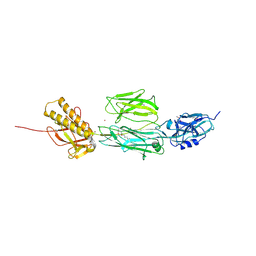 | | Full length structure of RrgB Pilus protein from Streptococcus pneumoniae | | Descriptor: | CELL WALL SURFACE ANCHOR FAMILY PROTEIN, NICKEL (II) ION | | Authors: | El-Mortaji, L, Contreras-Martel, C, Manzano, C, Vernet, T, Dessen, A, DiGuilmi, A.M. | | Deposit date: | 2010-12-10 | | Release date: | 2011-11-09 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | The Full-Length Streptococcus Pneumoniae Major Pilin Rrgb Crystallizes in a Fibre-Like Structure, which Presents the D1 Isopeptide Bond and Provides Details on the Mechanism of Pilus Polymerization.
Biochem.J., 441, 2012
|
|
2WJP
 
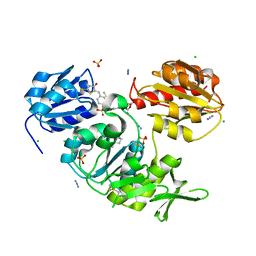 | | CRYSTAL STRUCTURE OF MURD LIGASE IN COMPLEX WITH D-GLU CONTAINING RHODANINE INHIBITOR | | Descriptor: | AZIDE ION, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Tomasic, T, Zidar, N, Sink, R, Kovac, A, Rupnik, V, Turk, S, Contreras-Martel, C, Dessen, A, Blanot, D, Muller-Premru, M, Gobec, S, Zega, A, Peterlin-Masic, L, Kikelj, D. | | Deposit date: | 2009-05-28 | | Release date: | 2010-08-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of Novel 5-Benzylidenerhodanine and 5-Benzylidenethiazolidine-2,4-Dione Inhibitors of Murd Ligase.
J.Med.Chem., 53, 2010
|
|
2X5O
 
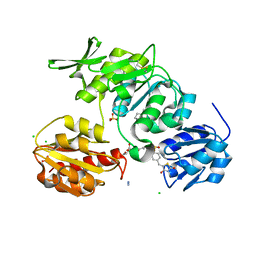 | | Discovery of Novel 5-Benzylidenerhodanine- and 5-Benzylidene- thiazolidine-2,4-dione Inhibitors of MurD Ligase | | Descriptor: | AZIDE ION, CHLORIDE ION, N-({3-[({4-[(Z)-(2,4-DIOXO-1,3-THIAZOLIDIN-5-YLIDENE)METHYL]PHENYL}AMINO)METHYL]PHENYL}CARBONYL)-D-GLUTAMIC ACID, ... | | Authors: | Zidar, N, Tomasic, T, Sink, R, Rupnik, V, Kovac, A, Turk, S, Contreras-Martel, C, Dessen, A, Blanot, D, Gobec, S, Zega, A, Peterlin-Masic, L, Kikelja, D. | | Deposit date: | 2010-02-10 | | Release date: | 2010-09-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Discovery of novel 5-benzylidenerhodanine and 5-benzylidenethiazolidine-2,4-dione inhibitors of MurD ligase.
J. Med. Chem., 53, 2010
|
|
2UWJ
 
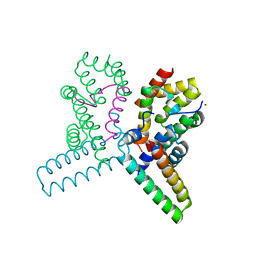 | | Structure of the heterotrimeric complex which regulates type III secretion needle formation | | Descriptor: | NICKEL (II) ION, TYPE III EXPORT PROTEIN PSCE, TYPE III EXPORT PROTEIN PSCF, ... | | Authors: | Quinaud, M, Ple, S, Job, V, Contreras-Martel, C, Simorre, J.P, Attree, I, Dessen, A. | | Deposit date: | 2007-03-22 | | Release date: | 2007-05-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the heterotrimeric complex that regulates type III secretion needle formation.
Proc. Natl. Acad. Sci. U.S.A., 104, 2007
|
|
2VTE
 
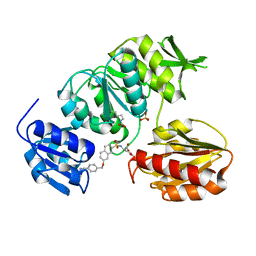 | | Crystal structure of MurD ligase in complex with D-Glu containing sulfonamide inhibitor | | Descriptor: | N-({7-[(4-cyanobenzyl)oxy]naphthalen-2-yl}sulfonyl)-D-glutamic acid, SULFATE ION, UDP-N-ACETYLMURAMOYLALANINE--D-GLUTAMATE LIGASE | | Authors: | Humljan, J, Kotnik, M, Contreras-Martel, C, Blanot, D, Urleb, U, Dessen, A, Solmajer, T, Gobec, S. | | Deposit date: | 2008-05-14 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Novel naphthalene-N-sulfonyl-D-glutamic acid derivatives as inhibitors of MurD, a key peptidoglycan biosynthesis enzyme.
J. Med. Chem., 51, 2008
|
|
2VTD
 
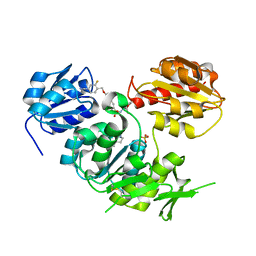 | | Crystal structure of MurD ligase in complex with D-Glu containing sulfonamide inhibitor | | Descriptor: | N-({6-[(4-CYANO-2-FLUOROBENZYL)OXY]NAPHTHALEN-2-YL}SULFONYL)-D-GLUTAMIC ACID, SULFATE ION, UDP-N-ACETYLMURAMOYLALANINE--D-GLUTAMATE LIGASE | | Authors: | Humljan, J, Kotnik, M, Contreras-Martel, C, Blanot, D, Urleb, U, Dessen, A, Solmajer, T, Gobec, S. | | Deposit date: | 2008-05-14 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Novel naphthalene-N-sulfonyl-D-glutamic acid derivatives as inhibitors of MurD, a key peptidoglycan biosynthesis enzyme.
J. Med. Chem., 51, 2008
|
|
2UUO
 
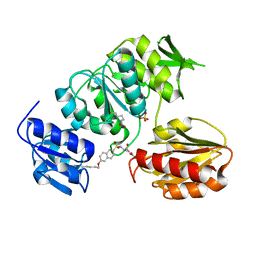 | | Crystal structure of MurD ligase in complex with D-Glu containing sulfonamide inhibitor | | Descriptor: | N-{[6-(PENTYLOXY)NAPHTHALEN-2-YL]SULFONYL}-D-GLUTAMIC ACID, SULFATE ION, UDP-N-ACETYLMURAMOYLALANINE--D-GLUTAMATE LIGASE | | Authors: | Humljan, J, Kotnik, M, Contreras-Martel, C, Blanot, D, Urleb, U, Dessen, A, Solmajer, T, Gobec, S. | | Deposit date: | 2007-03-06 | | Release date: | 2008-03-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Novel naphthalene-N-sulfonyl-D-glutamic acid derivatives as inhibitors of MurD, a key peptidoglycan biosynthesis enzyme.
J. Med. Chem., 51, 2008
|
|
2UUP
 
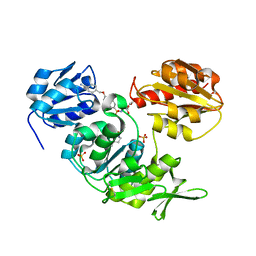 | | Crystal structure of MurD ligase in complex with D-Glu containing sulfonamide inhibitor | | Descriptor: | N-({6-[(4-CYANOBENZYL)OXY]NAPHTHALEN-2-YL}SULFONYL)-D-GLUTAMIC ACID, SULFATE ION, UDP-N-ACETYLMURAMOYLALANINE--D-GLUTAMATE LIGASE | | Authors: | Humljan, J, Kotnik, M, Contreras-Martel, C, Blanot, D, Urleb, U, Dessen, A, Solmajer, T, Gobec, S. | | Deposit date: | 2007-03-06 | | Release date: | 2008-03-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Novel naphthalene-N-sulfonyl-D-glutamic acid derivatives as inhibitors of MurD, a key peptidoglycan biosynthesis enzyme.
J. Med. Chem., 51, 2008
|
|
2VQ2
 
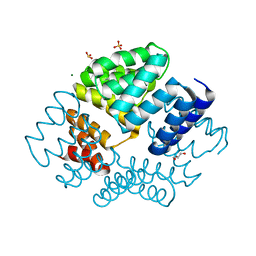 | | Crystal structure of PilW, widely conserved type IV pilus biogenesis factor | | Descriptor: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Trinidade, M.B, Job, V, Contreras-Martel, C, Pelicic, V, Dessen, A. | | Deposit date: | 2008-03-10 | | Release date: | 2008-05-27 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structure of a widely conserved type IV pilus biogenesis factor that affects the stability of secretin multimers.
J. Mol. Biol., 378, 2008
|
|
2Y2G
 
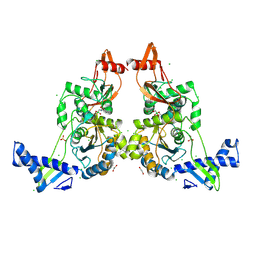 | | PENICILLIN-BINDING PROTEIN 1B (PBP-1B) IN COMPLEX WITH AN ALKYL BORONATE (A01) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, PENICILLIN-BINDING PROTEIN 1B, ... | | Authors: | Contreras-Martel, C, Amoroso, A, Woon, E.C, Zervosen, A, Inglis, S, Martins, A, Verlaine, O, Rydzik, A, Job, V, Luxen, A, Joris, B, Schofield, C.J, Dessen, A. | | Deposit date: | 2010-12-15 | | Release date: | 2011-08-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure-Guided Design of Cell Wall Biosynthesis Inhibitors that Overcome Beta-Lactam Resistance in Staphylococcus Aureus (Mrsa).
Acs Chem.Biol., 6, 2011
|
|
2Y2N
 
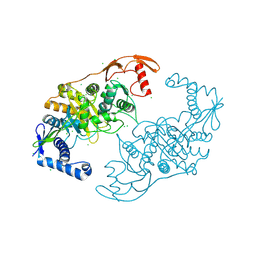 | | PENICILLIN-BINDING PROTEIN 1B (PBP-1B) IN COMPLEX WITH AN ALKYL BORONATE (E07) | | Descriptor: | CHLORIDE ION, PENICILLIN-BINDING PROTEIN 1B, SODIUM ION, ... | | Authors: | Contreras-Martel, C, Amoroso, A, Woon, E.C, Zervosen, A, Inglis, S, Martins, A, Verlaine, O, Rydzik, A, Job, V, Luxen, A, Joris, B, Schofield, C.J, Dessen, A. | | Deposit date: | 2010-12-15 | | Release date: | 2011-08-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structure-Guided Design of Cell Wall Biosynthesis Inhibitors that Overcome Beta-Lactam Resistance in Staphylococcus Aureus (Mrsa).
Acs Chem.Biol., 6, 2011
|
|
2Y2Q
 
 | | PENICILLIN-BINDING PROTEIN 1B (PBP-1B) IN COMPLEX WITH AN ALKYL BORONATE (Z06) | | Descriptor: | (3-acetamido-5-carboxy-phenyl)-trihydroxy-boron, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Contreras-Martel, C, Amoroso, A, Woon, E.C, Zervosen, A, Inglis, S, Martins, A, Verlaine, O, Rydzik, A, Job, V, Luxen, A, Joris, B, Schofield, C.J, Dessen, A. | | Deposit date: | 2010-12-15 | | Release date: | 2011-08-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure-Guided Design of Cell Wall Biosynthesis Inhibitors that Overcome Beta-Lactam Resistance in Staphylococcus Aureus (Mrsa).
Acs Chem.Biol., 6, 2011
|
|
2Y2O
 
 | | PENICILLIN-BINDING PROTEIN 1B (PBP-1B) IN COMPLEX WITH AN ALKYL BORONATE (EO9) | | Descriptor: | CHLORIDE ION, PENICILLIN-BINDING PROTEIN 1B, SODIUM ION, ... | | Authors: | Contreras-Martel, C, Amoroso, A, Woon, E.C, Zervosen, A, Inglis, S, Martins, A, Verlaine, O, Rydzik, A, Job, V, Luxen, A, Joris, B, Schofield, C.J, Dessen, A. | | Deposit date: | 2010-12-15 | | Release date: | 2011-08-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structure-Guided Design of Cell Wall Biosynthesis Inhibitors that Overcome Beta-Lactam Resistance in Staphylococcus Aureus (Mrsa).
Acs Chem.Biol., 6, 2011
|
|
2Y2I
 
 | | PENICILLIN-BINDING PROTEIN 1B (PBP-1B) IN COMPLEX WITH AN ALKYL BORONATE (ZA3) | | Descriptor: | CHLORIDE ION, PENICILLIN-BINDING PROTEIN 1B, SODIUM ION, ... | | Authors: | Contreras-Martel, C, Amoroso, A, Woon, E.C, Zervosen, A, Inglis, S, Martins, A, Verlaine, O, Rydzik, A, Job, V, Luxen, A, Joris, B, Schofield, C.J, Dessen, A. | | Deposit date: | 2010-12-15 | | Release date: | 2011-08-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure-Guided Design of Cell Wall Biosynthesis Inhibitors that Overcome Beta-Lactam Resistance in Staphylococcus Aureus (Mrsa).
Acs Chem.Biol., 6, 2011
|
|
4JMQ
 
 | | Crystal structure of pb9: The Dit of bacteriophage T5. | | Descriptor: | Bacteriophage T5 distal tail protein | | Authors: | Flayhan, A, Vellieux, F.M.D, Girard, E, Maury, O, Boulanger, P, Breyton, C. | | Deposit date: | 2013-03-14 | | Release date: | 2013-11-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.895 Å) | | Cite: | Crystal Structure of pb9, the Distal Tail Protein of Bacteriophage T5: a Conserved Structural Motif among All Siphophages.
J.Virol., 88, 2014
|
|
