6X7A
 
 | | Crystal structure of acetyltransferase Eis from Mycobacterium tuberculosis in complex with inhibitor SGT572 | | Descriptor: | 4-(4-cyclohexyl-3,4-dihydro-2~{H}-pyridin-1-yl)-1-(4-$l^{2}-fluoranylcyclohexa-1,3,5-trien-1-yl)butan-1-one, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Punetha, A, Garneau-Tsodikova, S, Tsodikov, O.V. | | Deposit date: | 2020-05-29 | | Release date: | 2021-06-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structure-based design of haloperidol analogues as inhibitors of acetyltransferase Eis from Mycobacterium tuberculosis to overcome kanamycin resistance
Rsc Med Chem, 12, 2021
|
|
3I6B
 
 | |
6OVQ
 
 | | Crystal structure of mithramycin 3-side chain keto-reductase MtmW | | Descriptor: | GLYCEROL, Putative Side chain reductase | | Authors: | Hou, C, Yu, X, Rohr, J, Tsodikov, O.V. | | Deposit date: | 2019-05-08 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of a Cryptic Intermediate in Late Steps of Mithramycin Biosynthesis.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6OVX
 
 | | Crystal structure of mithramycin 3-side chain keto-reductase MtmW in complex with NAD+, P422 form | | Descriptor: | GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Putative side chain reductase | | Authors: | Hou, C, Yu, X, Rohr, J, Tsodikov, O.V. | | Deposit date: | 2019-05-08 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of a Cryptic Intermediate in Late Steps of Mithramycin Biosynthesis.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6OW0
 
 | | Crystal structure of mithramycin 3-side chain keto-reductase MtmW in complex with NAD+ and PEG | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, MtmW, ... | | Authors: | Hou, C, Yu, X, Rohr, J, Tsodikov, O.V. | | Deposit date: | 2019-05-08 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Discovery of a Cryptic Intermediate in Late Steps of Mithramycin Biosynthesis.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
3K6Z
 
 | | Crystal structure of Rv3671c protease, inactive form | | Descriptor: | POSSIBLE MEMBRANE-ASSOCIATED SERINE PROTEASE | | Authors: | Biswas, T, Small, J, Vandal, O, Ehrt, S, Tsodikov, O.V. | | Deposit date: | 2009-10-10 | | Release date: | 2010-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural insight into serine protease Rv3671c that Protects M. tuberculosis from oxidative and acidic stress.
Structure, 18, 2010
|
|
5DBJ
 
 | | Crystal structure of halogenase PltA | | Descriptor: | CHLORIDE ION, FADH2-dependent halogenase PltA, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Pang, A.H, Tsodikov, O.V. | | Deposit date: | 2015-08-21 | | Release date: | 2015-10-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure of halogenase PltA from the pyoluteorin biosynthetic pathway.
J.Struct.Biol., 192, 2015
|
|
9BDW
 
 | | NF-kappaB RelA homo-dimer bound to GC-centric kappaB DNA | | Descriptor: | DNA (5'-D(P*AP*CP*TP*GP*GP*GP*AP*AP*GP*TP*TP*CP*CP*AP*GP*TP*GP*AP*T)-3'), DNA (5'-D(P*AP*TP*CP*AP*CP*TP*GP*GP*AP*AP*CP*TP*TP*CP*CP*CP*AP*GP*T)-3'), SULFATE ION, ... | | Authors: | Biswas, T, Shahabi, S, Tsodikov, O.V, Huang, D, Ghosh, G. | | Deposit date: | 2024-04-12 | | Release date: | 2024-04-24 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Transient interactions modulate the affinity of NF-kappa B transcription factors for DNA.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
9BDX
 
 | | NF-kappaB RelA homo-dimer bound to CG-centric kappaB DNA | | Descriptor: | DNA (5'-D(*AP*TP*CP*AP*CP*TP*GP*GP*AP*AP*GP*TP*TP*CP*CP*CP*AP*GP*T)-3'), DNA (5'-D(P*AP*CP*TP*GP*GP*GP*AP*AP*CP*TP*TP*CP*CP*AP*GP*TP*GP*AP*T)-3'), Transcription factor p65 | | Authors: | Biswas, T, Shahabi, S, Tsodikov, O.V, Ghosh, G. | | Deposit date: | 2024-04-12 | | Release date: | 2024-04-24 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Transient interactions modulate the affinity of NF-kappa B transcription factors for DNA.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
9BDV
 
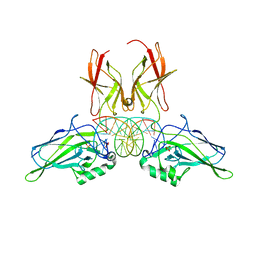 | | NF-kappaB RelA homo-dimer bound to TA-centric kappaB DNA | | Descriptor: | DNA (5'-D(*AP*TP*CP*AP*CP*TP*GP*GP*AP*AP*AP*TP*TP*CP*CP*CP*AP*GP*T)-3'), DNA (5'-D(P*AP*CP*TP*GP*GP*GP*AP*AP*TP*TP*TP*CP*CP*AP*GP*TP*GP*AP*T)-3'), SULFATE ION, ... | | Authors: | Biswas, T, Shahabi, S, Tsodikov, O.V. | | Deposit date: | 2024-04-12 | | Release date: | 2024-04-24 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Transient interactions modulate the affinity of NF-kappa B transcription factors for DNA.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
5TVJ
 
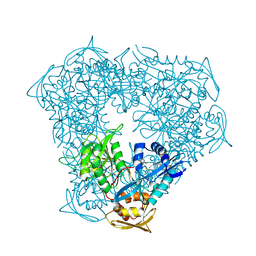 | | Crystal structure of acetyltransferase Eis from Mycobacterium tuberculosis in complex with CoA and inhibitor 2k*: 1-(4-fluorophenyl)-2-[2-(4-methylphenyl)-2-oxoethyl]pyrrolo[1,2-a]pyrazin-2-ium | | Descriptor: | 1-(4-fluorophenyl)-2-[2-(4-methylphenyl)-2-oxoethyl]pyrrolo[1,2-a]pyrazin-2-ium, CHLORIDE ION, COENZYME A, ... | | Authors: | Gajadeera, C.S, Garzan, A, Hou, C, Garneau-Tsodikova, S, Tsodikov, O.V. | | Deposit date: | 2016-11-09 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Combating Enhanced Intracellular Survival (Eis)-Mediated Kanamycin Resistance of Mycobacterium tuberculosis by Novel Pyrrolo[1,5-a]pyrazine-Based Eis Inhibitors.
ACS Infect Dis, 3, 2017
|
|
4I9F
 
 | | Crystal structure of glycerol phosphate phosphatase Rv1692 from Mycobacterium tuberculosis in complex with calcium | | Descriptor: | CALCIUM ION, CHLORIDE ION, Glycerol 3-phosphate phosphatase | | Authors: | Biswas, T, Larrouy-Maumus, G, de Carvalho, L.P, Tsodikov, O.V. | | Deposit date: | 2012-12-05 | | Release date: | 2013-07-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Discovery of a glycerol 3-phosphate phosphatase reveals glycerophospholipid polar head recycling in Mycobacterium tuberculosis.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
5KDE
 
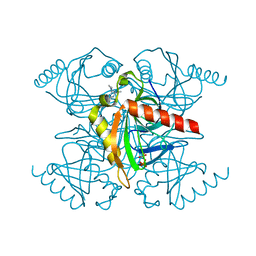 | | Inorganic pyrophosphatase from Mycobacterium tuberculosis in complex with inhibitor 1 and inorganic pyrophosphate | | Descriptor: | 2,4-bis(aziridin-1-yl)-6-(1-phenylpyrrol-2-yl)-1,3,5-triazine, Inorganic pyrophosphatase, PYROPHOSPHATE 2- | | Authors: | Pang, A.H, Garzan, A, Garneau-Tsodikova, S, Tsodikov, O.V. | | Deposit date: | 2016-06-08 | | Release date: | 2016-09-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Discovery of Allosteric and Selective Inhibitors of Inorganic Pyrophosphatase from Mycobacterium tuberculosis.
ACS Chem. Biol., 11, 2016
|
|
5KDF
 
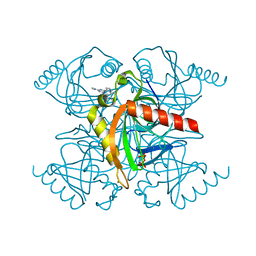 | | Inorganic pyrophosphatase from Mycobacterium tuberculosis in complex with inhibitor 6 and inorganic pyrophosphate | | Descriptor: | CALCIUM ION, Inorganic pyrophosphatase, PYROPHOSPHATE 2-, ... | | Authors: | Pang, A.H, Garzan, A, Garneau-Tsodikova, S, Tsodikov, O.V. | | Deposit date: | 2016-06-08 | | Release date: | 2016-09-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Discovery of Allosteric and Selective Inhibitors of Inorganic Pyrophosphatase from Mycobacterium tuberculosis.
ACS Chem. Biol., 11, 2016
|
|
4IJJ
 
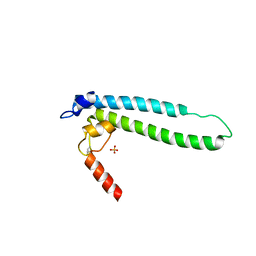 | | Structure of transcription factor DksA2 from Pseudomonas aeruginosa | | Descriptor: | Putative C4-type zinc finger protein, DksA/TraR family, SULFATE ION | | Authors: | Biswas, T, Furman, R, Artsimovitch, I, Tsodikov, O.V. | | Deposit date: | 2012-12-21 | | Release date: | 2013-02-27 | | Last modified: | 2013-03-27 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | DksA2, a zinc-independent structural analog of the transcription factor DksA.
Febs Lett., 587, 2013
|
|
5W35
 
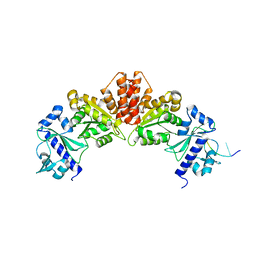 | |
5W36
 
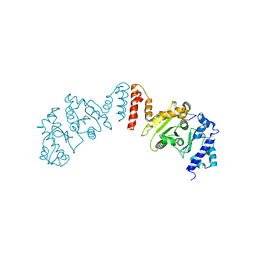 | |
6DXJ
 
 | |
6DY5
 
 | |
5WHF
 
 | |
5W33
 
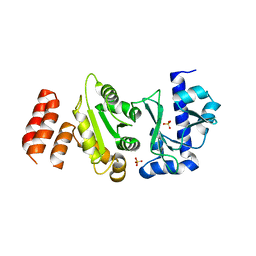 | |
5W34
 
 | |
6DWT
 
 | |
8D1R
 
 | | Crystal structure of acetyltransferase Eis from Mycobacterium tuberculosis in complex with inhibitor SGT520 | | Descriptor: | 1-{3-[(4-chlorophenyl)methoxy]phenyl}methanamine, GLYCEROL, N-acetyltransferase Eis | | Authors: | Pang, A.H, Punetha, A, Garneau-Tsodikova, S, Tsodikov, O.V. | | Deposit date: | 2022-05-27 | | Release date: | 2022-09-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Discovery of substituted benzyloxy-benzylamine inhibitors of acetyltransferase Eis and their anti-mycobacterial activity.
Eur.J.Med.Chem., 242, 2022
|
|
8D25
 
 | | Crystal structure of acetyltransferase Eis from Mycobacterium tuberculosis in complex with inhibitor SGT530 | | Descriptor: | 1-{2-[(3-chlorophenyl)methoxy]phenyl}-N-[(pyridin-4-yl)methyl]methanamine, GLYCEROL, N-acetyltransferase Eis | | Authors: | Pang, A.H, Punetha, A, Garneau-Tsodikova, S, Tsodikov, O.V. | | Deposit date: | 2022-05-27 | | Release date: | 2022-09-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery of substituted benzyloxy-benzylamine inhibitors of acetyltransferase Eis and their anti-mycobacterial activity.
Eur.J.Med.Chem., 242, 2022
|
|
