7YY7
 
 | |
7YY8
 
 | |
7YY4
 
 | |
7YYA
 
 | |
7YXZ
 
 | |
7YWT
 
 | | human Carbonic Anhydrase II in complex with 4-oxo-N-(4-sulfamoylphenyl)-1,3,4,6,7,11b-hexahydro-2H-pyrazino[2,1-a]isoquinoline-2-carboxamide | | Descriptor: | (11~{b}~{R})-4-oxidanylidene-~{N}-(4-sulfamoylphenyl)-3,6,7,11~{b}-tetrahydro-1~{H}-pyrazino[2,1-a]isoquinoline-2-carboxamide, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Angeli, A, Ferraroni, M. | | Deposit date: | 2022-02-14 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.106 Å) | | Cite: | Development of Praziquantel sulphonamide derivatives as antischistosomal drugs.
J Enzyme Inhib Med Chem, 37, 2022
|
|
7YZH
 
 | | Schistosoma Mansoni Carbonic Anhydrase in complex with 4-oxo-N-(4-sulfamoylphenethyl)-1,3,4,6,7,11b-hexahydro-2H-pyrazino[2,1-a]isoquinoline-2-carbothioamide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-oxo-N-(4-sulfamoylphenethyl)-1,3,4,6,7,11b-hexahydro-2H-pyrazino[2,1-a]isoquinoline-2-carbothioamide, Carbonic anhydrase, ... | | Authors: | Angeli, A, Ferraroni, M. | | Deposit date: | 2022-02-20 | | Release date: | 2023-03-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Development of Praziquantel sulphonamide derivatives as antischistosomal drugs.
J Enzyme Inhib Med Chem, 37, 2022
|
|
5UDE
 
 | | Crystal Structure of RSV F B9320 DS-Cav1 | | Descriptor: | Fusion glycoprotein F0, SULFATE ION | | Authors: | McLellan, J.S. | | Deposit date: | 2016-12-26 | | Release date: | 2017-05-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | A highly potent extended half-life antibody as a potential RSV vaccine surrogate for all infants.
Sci Transl Med, 9, 2017
|
|
6QMH
 
 | |
6QMG
 
 | |
6QMI
 
 | |
6QMF
 
 | |
8OFI
 
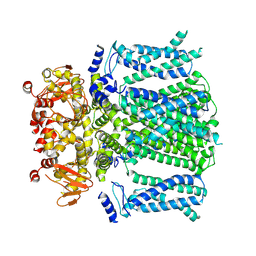 | | Ivabradine bound to HCN4 channel | | Descriptor: | Ivabradine, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 | | Authors: | Saponaro, A, Chaves-Sanjuan, A, Sharifzadeh, A.S, Clarke, O.B, Marabelli, C, Bolognesi, M, Thiel, G, Moroni, A. | | Deposit date: | 2023-03-15 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural determinants of ivabradine block of the open pore of HCN4.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
5I04
 
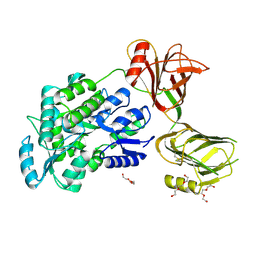 | | Crystal structure of the orphan region of human endoglin/CD105 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Maltose-binding periplasmic protein,Endoglin, TRIETHYLENE GLYCOL, ... | | Authors: | Saito, T, Bokhove, M, de Sanctis, D, Jovine, L. | | Deposit date: | 2016-02-03 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural Basis of the Human Endoglin-BMP9 Interaction: Insights into BMP Signaling and HHT1.
Cell Rep, 19, 2017
|
|
3KMB
 
 | |
5JZD
 
 | | A re-refinement of the isochorismate synthase EntC | | Descriptor: | (5S,6S)-5-[(1-carboxyethenyl)oxy]-6-hydroxycyclohexa-1,3-diene-1-carboxylic acid, Isochorismate synthase EntC, MAGNESIUM ION | | Authors: | Meneely, K.M, Lamb, A.L. | | Deposit date: | 2016-05-16 | | Release date: | 2016-07-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | An Open and Shut Case: The Interaction of Magnesium with MST Enzymes.
J.Am.Chem.Soc., 138, 2016
|
|
2IVZ
 
 | | Structure of TolB in complex with a peptide of the colicin E9 T- domain | | Descriptor: | CALCIUM ION, COLICIN-E9, PROTEIN TOLB | | Authors: | Loftus, S.R, Walker, D, Mate, M.J, Bonsor, D.A, James, R, Moore, G.R, Kleanthous, C. | | Deposit date: | 2006-06-23 | | Release date: | 2006-08-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Competitive Recruitment of the Periplasmic Translocation Portal Tolb by a Natively Disordered Domain of Colicin E9
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
5JFG
 
 | | Structure of humanised RadA-mutant humRadA22F in complex with peptide FHTA | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, DNA repair and recombination protein RadA, ... | | Authors: | Fischer, G, Marsh, M, Moschetti, T, Sharpe, T, Scott, D, Morgan, M, Ng, H, Skidmore, J, Venkitaraman, A, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2016-04-19 | | Release date: | 2016-10-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Humanisation of RadA from Pyrococcus furiosus
To Be Published
|
|
4KMB
 
 | |
5HZW
 
 | | Crystal structure of the orphan region of human endoglin/CD105 in complex with BMP9 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Growth/differentiation factor 2, Maltose-binding periplasmic protein,Endoglin, ... | | Authors: | Bokhove, M, Saito, T, Jovine, L. | | Deposit date: | 2016-02-03 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (4.451 Å) | | Cite: | Structural Basis of the Human Endoglin-BMP9 Interaction: Insights into BMP Signaling and HHT1.
Cell Rep, 19, 2017
|
|
1B6F
 
 | | BIRCH POLLEN ALLERGEN BET V 1 | | Descriptor: | PROTEIN (MAJOR POLLEN ALLERGEN BET V 1-A) | | Authors: | Schweimer, K, Sticht, H, Boehm, M, Roesch, P. | | Deposit date: | 1999-01-13 | | Release date: | 2000-01-17 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR Spectroscopy Reveals Common Structural Features of the Birch Pollen Allergen Bet v 1 and the cherry allergen Pru a 1
APPL.MAGN.RESON., 17, 1999
|
|
6MJY
 
 | |
6NMC
 
 | | CryoEM structure of the LbCas12a-crRNA-2xAcrVA1 complex | | Descriptor: | AcrVA1, Cpf1, MAGNESIUM ION, ... | | Authors: | Chang, L, Li, Z, Zhang, H. | | Deposit date: | 2019-01-10 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.24 Å) | | Cite: | Structural Basis for the Inhibition of CRISPR-Cas12a by Anti-CRISPR Proteins.
Cell Host Microbe, 25, 2019
|
|
2NAO
 
 | | Atomic resolution structure of a disease-relevant Abeta(1-42) amyloid fibril | | Descriptor: | Beta-amyloid protein 42 | | Authors: | Waelti, M.A, Ravotti, F, Arai, H, Glabe, C, Wall, J, Bockmann, A, Guntert, P, Meier, B.H, Riek, R. | | Deposit date: | 2016-01-07 | | Release date: | 2016-07-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Atomic-resolution structure of a disease-relevant A beta (1-42) amyloid fibril.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5OU1
 
 | | M. thermoresistible IMPDH in complex with IMP and Compound 1 (7759844) | | Descriptor: | (2~{S})-~{N}-(4-iodophenyl)-2-(4-methoxyphenoxy)propanamide, INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase,Inosine-5'-monophosphate dehydrogenase | | Authors: | Ascher, D.B, Pacitto, A, Blundell, T.L. | | Deposit date: | 2017-08-23 | | Release date: | 2018-03-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Fragment-Based Approach to Targeting Inosine-5'-monophosphate Dehydrogenase (IMPDH) from Mycobacterium tuberculosis.
J. Med. Chem., 61, 2018
|
|
