3FDE
 
 | | Mouse UHRF1 SRA domain bound with hemi-methylated CpG DNA, crystal structure in space group C222(1) at 1.4 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 5'-D(*CP*CP*AP*TP*GP*(5CM)P*GP*CP*TP*GP*AP*C)-3', 5'-D(*GP*TP*CP*AP*GP*CP*GP*CP*AP*TP*GP*G)-3', ... | | Authors: | Hashimoto, H, Horton, J.R, Zhang, X, Cheng, X. | | Deposit date: | 2008-11-25 | | Release date: | 2009-01-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | UHRF1, a modular multi-domain protein, regulates replication-coupled crosstalk between DNA methylation and histone modifications.
Epigenetics, 4, 2009
|
|
3F8J
 
 | | Mouse UHRF1 SRA domain bound with hemi-methylated CpG, crystal structure in space group C222(1) | | Descriptor: | 5'-D(*DCP*DCP*DAP*DTP*DGP*(5CM)P*DGP*DCP*DTP*DGP*DAP*DC)-3', 5'-D(*DGP*DTP*DCP*DAP*DGP*DCP*DGP*DCP*DAP*DTP*DGP*DG)-3', E3 ubiquitin-protein ligase UHRF1, ... | | Authors: | Hashimoto, H, Horton, J.R, Zhang, X, Cheng, X. | | Deposit date: | 2008-11-12 | | Release date: | 2009-01-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | UHRF1, a modular multi-domain protein, regulates replication-coupled crosstalk between DNA methylation and histone modifications.
Epigenetics, 4, 2009
|
|
2L6K
 
 | | Solution Structure of a Nonphosphorylated Peptide Recognizing Domain | | Descriptor: | Tensin-like C1 domain-containing phosphatase | | Authors: | Dai, K, Liao, S, Zhang, J, Zhang, X, Tu, X. | | Deposit date: | 2010-11-22 | | Release date: | 2011-10-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Tensin2 SH2 domain and its phosphotyrosine-independent interaction with DLC-1
Plos One, 6, 2011
|
|
6LZ3
 
 | |
5YZG
 
 | | The Cryo-EM Structure of Human Catalytic Step I Spliceosome (C complex) at 4.1 angstrom resolution | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Zhan, X, Yan, C, Zhang, X, Lei, J, Shi, Y. | | Deposit date: | 2017-12-14 | | Release date: | 2018-08-08 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structure of a human catalytic step I spliceosome
Science, 359, 2018
|
|
6AH0
 
 | | The Cryo-EM Structure of the Precusor of Human Pre-catalytic Spliceosome (pre-B complex) | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Zhan, X, Yan, C, Zhang, X, Shi, Y. | | Deposit date: | 2018-08-15 | | Release date: | 2018-11-14 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Structures of the human pre-catalytic spliceosome and its precursor spliceosome.
Cell Res., 28, 2018
|
|
6AHD
 
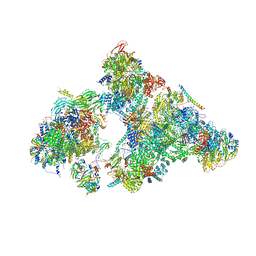 | | The Cryo-EM Structure of Human Pre-catalytic Spliceosome (B complex) at 3.8 angstrom resolution | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, Brr2, U5 small nuclear ribonucleoprotein 200 kDa helicase, ... | | Authors: | Zhan, X, Yan, C, Zhang, X, Shi, Y. | | Deposit date: | 2018-08-17 | | Release date: | 2018-11-14 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structures of the human pre-catalytic spliceosome and its precursor spliceosome.
Cell Res., 28, 2018
|
|
5YF0
 
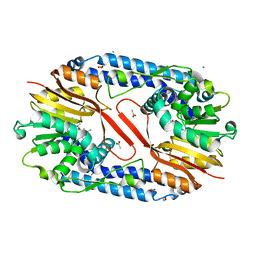 | | Crystal structure of CARNMT1 bound to SAM | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Cao, R, Zhang, X, Li, H. | | Deposit date: | 2017-09-20 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Molecular basis for histidine N1 position-specific methylation by CARNMT1.
Cell Res., 28, 2018
|
|
6S8F
 
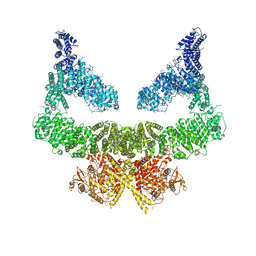 | | Structure of nucleotide-bound Tel1/ATM | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Serine/threonine-protein kinase TEL1,Serine/threonine-protein kinase TEL1,Serine/threonine-protein kinase TEL1,Serine/threonine-protein kinase TEL1,Serine/threonine-protein kinase TEL1 | | Authors: | Yates, L.A, Williams, R.M, Ayala, R, Zhang, X. | | Deposit date: | 2019-07-09 | | Release date: | 2019-10-30 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM Structure of Nucleotide-Bound Tel1ATMUnravels the Molecular Basis of Inhibition and Structural Rationale for Disease-Associated Mutations.
Structure, 28, 2020
|
|
5EZK
 
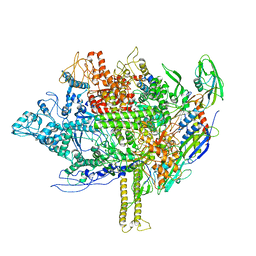 | | RNA polymerase model placed by Molecular replacement into X-ray diffraction map of DNA-bound RNA Polymerase-Sigma 54 holoenzyme complex. | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Darbari, V.C, Yang, Y, Lu, D, Zhang, N, Glyde, R, Wang, Y, Murakami, K.S, Buck, M, Zhang, X. | | Deposit date: | 2015-11-26 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (8.5 Å) | | Cite: | TRANSCRIPTION. Structures of the RNA polymerase- Sigma 54 reveal new and conserved regulatory strategies.
Science, 349, 2015
|
|
7EU3
 
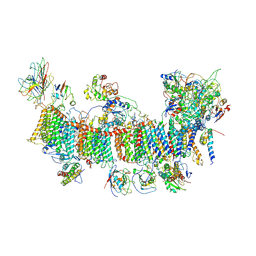 | | Chloroplast NDH complex | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, BETA-CAROTENE, ... | | Authors: | Wang, W.D, Shen, L, Tang, K, Han, G.Y, Zhang, X, Shen, J.R. | | Deposit date: | 2021-05-15 | | Release date: | 2021-12-29 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Architecture of the chloroplast PSI-NDH supercomplex in Hordeum vulgare.
Nature, 601, 2022
|
|
7EWK
 
 | | Barley photosystem I-LHCI-Lhca6 supercomplex | | Descriptor: | (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Wang, W.D, Shen, L, Tang, K, Han, G.Y, Zhang, X, Shen, J.R. | | Deposit date: | 2021-05-25 | | Release date: | 2022-01-12 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.88 Å) | | Cite: | Architecture of the chloroplast PSI-NDH supercomplex in Hordeum vulgare.
Nature, 601, 2022
|
|
9JF4
 
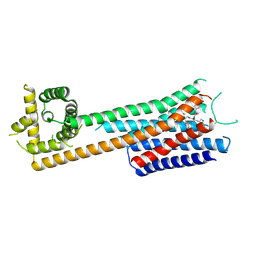 | | cryo-EM structure of Neuromedin B receptor in complex with PD168368 | | Descriptor: | (2~{S})-3-(1~{H}-indol-3-yl)-2-methyl-2-[(4-nitrophenyl)carbamoylamino]-~{N}-[(1-pyridin-2-ylcyclohexyl)methyl]propanamide, Neuromedin-B receptor,de novo design protein | | Authors: | Gao, K, Zhang, X, Liu, X. | | Deposit date: | 2024-09-03 | | Release date: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | cryo-EM structure of Neuromedin B receptor in complex with PD168368
To Be Published
|
|
3EA3
 
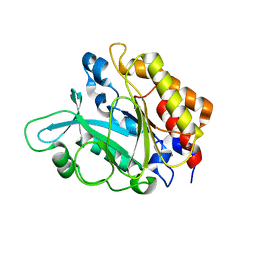 | | Crystal Structure of the Y246S/Y247S/Y248S/Y251S Mutant of Phosphatidylinositol-Specific Phospholipase C from Bacillus Thuringiensis | | Descriptor: | 1-phosphatidylinositol phosphodiesterase, MANGANESE (II) ION | | Authors: | Shi, X, Shao, C, Zhang, X, Zambonelli, C, Redfied, A.G, Head, J.F, Seaton, B.A, Roberts, M.F. | | Deposit date: | 2008-08-24 | | Release date: | 2009-04-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Modulation of Bacillus thuringiensis Phosphatidylinositol-specific Phospholipase C Activity by Mutations in the Putative Dimerization Interface.
J.Biol.Chem., 284, 2009
|
|
3EA2
 
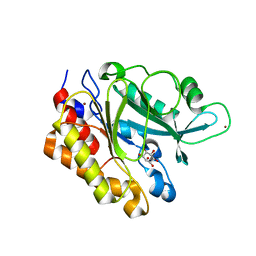 | | Crystal Structure of the Myo-inositol bound Y247S/Y251S Mutant of Phosphatidylinositol-Specific Phospholipase C from Bacillus Thuringiensis | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, 1-phosphatidylinositol phosphodiesterase, ZINC ION | | Authors: | Shi, X, Shao, C, Zhang, X, Zambonelli, C, Redfied, A.G, Head, J.F, Seaton, B.A, Roberts, M.F. | | Deposit date: | 2008-08-24 | | Release date: | 2009-04-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Modulation of bacillus thuringiensis phosphatidylinositol-specific phospholipase C activity by mutations in the putative dimerization interface.
J.Biol.Chem., 284, 2009
|
|
3EA1
 
 | | Crystal Structure of the Y247S/Y251S Mutant of Phosphatidylinositol-Specific Phospholipase C from Bacillus Thuringiensis | | Descriptor: | 1-phosphatidylinositol phosphodiesterase, ZINC ION | | Authors: | Shi, X, Shao, C, Zhang, X, Zambonelli, C, Redfied, A.G, Head, J.F, Seaton, B.A, Roberts, M.F. | | Deposit date: | 2008-08-24 | | Release date: | 2009-04-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Modulation of bacillus thuringiensis phosphatidylinositol-specific phospholipase C activity by mutations in the putative dimerization interface.
J.Biol.Chem., 284, 2009
|
|
3FPD
 
 | | G9a-like protein lysine methyltransferase inhibition by BIX-01294 | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-9 specific 5, N-(1-benzylpiperidin-4-yl)-6,7-dimethoxy-2-(4-methyl-1,4-diazepan-1-yl)quinazolin-4-amine, ... | | Authors: | Chang, Y, Zhang, X, Horton, J.R, Cheng, X. | | Deposit date: | 2009-01-05 | | Release date: | 2009-02-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for G9a-like protein lysine methyltransferase inhibition by BIX-01294.
Nat.Struct.Mol.Biol., 16, 2009
|
|
1AWP
 
 | | RAT OUTER MITOCHONDRIAL MEMBRANE CYTOCHROME B5 | | Descriptor: | CYTOCHROME B5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Wang, X, Zhang, X. | | Deposit date: | 1997-10-03 | | Release date: | 1998-10-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The reduction potential of cytochrome b5 is modulated by its exposed heme edge.
Biochemistry, 37, 1998
|
|
8BNZ
 
 | | BAM-EspP complex structure with BamA-G431C/EspP-N1293C mutations in nanodisc | | Descriptor: | Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, Outer membrane protein assembly factor BamC, ... | | Authors: | Shen, C, Chang, S, Luo, Q, Zhang, Z, Xie, T, Luo, B, Lu, G, Zhu, X, Wei, X, Dong, C, Zhou, R, Zhang, X, Tang, X, Dong, H. | | Deposit date: | 2022-11-14 | | Release date: | 2023-04-26 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of BAM-mediated outer membrane beta-barrel protein assembly.
Nature, 617, 2023
|
|
8BO2
 
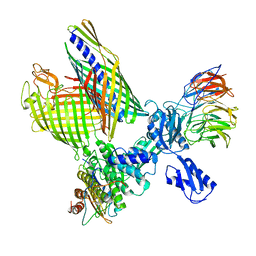 | | BAM-EspP complex structure with BamA-S425C/EspP-S1299C mutations in nanodisc | | Descriptor: | Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, Outer membrane protein assembly factor BamC, ... | | Authors: | Shen, C, Chang, S, Luo, Q, Zhang, Z, Xie, T, Luo, B, Lu, G, Zhu, X, Wei, X, Dong, C, Zhou, R, Zhang, X, Tang, X, Dong, H. | | Deposit date: | 2022-11-14 | | Release date: | 2023-04-26 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of BAM-mediated outer membrane beta-barrel protein assembly.
Nature, 617, 2023
|
|
8EQB
 
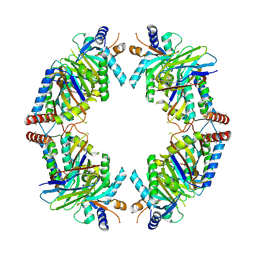 | | FAM46C/BCCIPalpha/Nanobody complex | | Descriptor: | Isoform 2 of BRCA2 and CDKN1A-interacting protein, Synthetic nanobody 1, Terminal nucleotidyltransferase 5C | | Authors: | Liu, S, Chen, H, Yin, Y, Bai, X, Zhang, X. | | Deposit date: | 2022-10-07 | | Release date: | 2023-03-15 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Inhibition of FAM46/TENT5 activity by BCCIP alpha adopting a unique fold.
Sci Adv, 9, 2023
|
|
6NT9
 
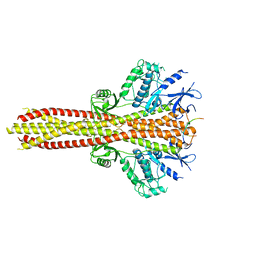 | | Cryo-EM structure of the complex between human TBK1 and chicken STING | | Descriptor: | Serine/threonine-protein kinase TBK1, Stimulator of interferon genes protein | | Authors: | Shang, G, Zhang, C, Chen, Z.J, Bai, X, Zhang, X. | | Deposit date: | 2019-01-28 | | Release date: | 2019-03-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of STING binding with and phosphorylation by TBK1.
Nature, 567, 2019
|
|
9J35
 
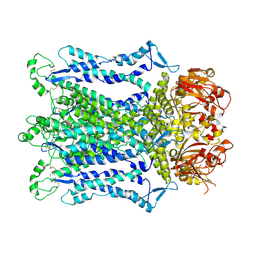 | | Cryo-EM structure of Arabidopsis CNGC5 in nanodisc | | Descriptor: | Probable cyclic nucleotide-gated ion channel 5 | | Authors: | Wang, J.P, Zhang, X, Zhang, P. | | Deposit date: | 2024-08-07 | | Release date: | 2025-02-19 | | Last modified: | 2025-04-02 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Cryo-EM structures of Arabidopsis CNGC1 and CNGC5 reveal molecular mechanisms underlying gating and calcium selectivity.
Nat.Plants, 11, 2025
|
|
9J36
 
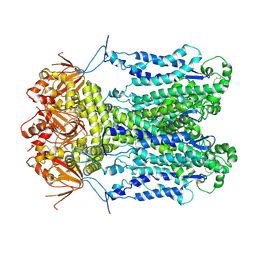 | | Cryo-EM structure of Arabidopsis CNGC5 | | Descriptor: | Probable cyclic nucleotide-gated ion channel 5 | | Authors: | Wang, J.P, Zhang, P, Zhang, X. | | Deposit date: | 2024-08-07 | | Release date: | 2025-02-19 | | Last modified: | 2025-04-02 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Cryo-EM structures of Arabidopsis CNGC1 and CNGC5 reveal molecular mechanisms underlying gating and calcium selectivity.
Nat.Plants, 11, 2025
|
|
9J34
 
 | | Cryo-EM structure of Arabidopsis CNGC1 | | Descriptor: | CALCIUM ION, Cyclic nucleotide-gated ion channel 1 | | Authors: | Wang, J.P, Zhang, P, Zhang, X. | | Deposit date: | 2024-08-07 | | Release date: | 2025-02-19 | | Last modified: | 2025-04-02 | | Method: | ELECTRON MICROSCOPY (2.51 Å) | | Cite: | Cryo-EM structures of Arabidopsis CNGC1 and CNGC5 reveal molecular mechanisms underlying gating and calcium selectivity.
Nat.Plants, 11, 2025
|
|
