7D5Z
 
 | | Crystal structure of EBV gH/gL bound with neutralizing antibody 1D8 | | Descriptor: | Envelope glycoprotein H, Envelope glycoprotein L, heavy chain of 1D8, ... | | Authors: | Zhu, Q, Shan, S, Yu, J, Wang, X, Zhang, L, Zeng, M. | | Deposit date: | 2020-09-28 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | A Neutralizing Antibody Targeting a New Site of Vulnerability on Epstein-Barr Virus gH/gL Protects against Dual-Tropic Infection
To Be Published
|
|
7YAN
 
 | |
7W5G
 
 | | The apo structure of trichobrasilenol synthase TaTC6 with the space group of orthorhombic | | Descriptor: | GLYCEROL, SULFATE ION, Terpene cyclase 6 | | Authors: | Chen, C, Wang, T, Yang, Y, Zhang, L, Ko, T, Huang, J, Guo, R. | | Deposit date: | 2021-11-30 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into the cyclization of unusual brasilane-type sesquiterpenes.
Int.J.Biol.Macromol., 209, 2022
|
|
7W5F
 
 | | The apo structure of trichobrasilenol synthase TaTC6 with the space group of monoclinic | | Descriptor: | MAGNESIUM ION, Terpene cyclase 6 | | Authors: | Chen, C, Wang, T, Yang, Y, Zhang, L, Ko, T, Huang, J, Guo, R. | | Deposit date: | 2021-11-30 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structural insights into the cyclization of unusual brasilane-type sesquiterpenes.
Int.J.Biol.Macromol., 209, 2022
|
|
7W5I
 
 | | The structure of trichobrasilenol synthase TaTC6 in complex with FPP-1 | | Descriptor: | FARNESYL DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Chen, C, Wang, T, Yang, Y, Zhang, L, Ko, T, Huang, J, Guo, R. | | Deposit date: | 2021-11-30 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structural insights into the cyclization of unusual brasilane-type sesquiterpenes.
Int.J.Biol.Macromol., 209, 2022
|
|
7CK6
 
 | | Protein translocase of mitochondria | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Mitochondrial import receptor subunit TOM22 homolog, Mitochondrial import receptor subunit TOM40 homolog, ... | | Authors: | Yang, M, Wang, W, Zhang, L, Chen, X. | | Deposit date: | 2020-07-15 | | Release date: | 2020-11-04 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Atomic structure of human TOM core complex.
Cell Discov, 6, 2020
|
|
6O3G
 
 | |
7NZN
 
 | | Structure of RET kinase domain bound to inhibitor JB-48 | | Descriptor: | 2-[4-[[4-[1-[2-(dimethylamino)ethyl]pyrazol-4-yl]-6-[(3-methyl-1~{H}-pyrazol-5-yl)amino]pyrimidin-2-yl]amino]phenyl]-~{N}-(3-fluorophenyl)ethanamide, FORMIC ACID, Proto-oncogene tyrosine-protein kinase receptor Ret | | Authors: | Briggs, D.C, McDonald, N.Q. | | Deposit date: | 2021-03-24 | | Release date: | 2022-02-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Discovery of N-Trisubstituted Pyrimidine Derivatives as Type I RET and RET Gatekeeper Mutant Inhibitors with a Novel Kinase Binding Pose.
J.Med.Chem., 65, 2022
|
|
6LDL
 
 | | Crystal structure of CYP116B46-N(20-445) from Tepidiphilus thermophilus in complex with HEME | | Descriptor: | BICINE, Cytochrome P450, GLYCEROL, ... | | Authors: | Zhang, L.L, Xie, Z.Z, Huang, J.W, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2019-11-21 | | Release date: | 2020-10-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Structural insight into the electron transfer pathway of a self-sufficient P450 monooxygenase.
Nat Commun, 11, 2020
|
|
2YHN
 
 | | The IDOL-UBE2D complex mediates sterol-dependent degradation of the LDL receptor | | Descriptor: | E3 UBIQUITIN-PROTEIN LIGASE MYLIP, ZINC ION | | Authors: | Fairall, L, Goult, B.T, Millard, C.J, Tontonoz, P, Schwabe, J.W.R. | | Deposit date: | 2011-05-04 | | Release date: | 2011-06-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Idol-Ube2D Complex Mediates Sterol-Dependent Degradation of the Ldl Receptor.
Genes Dev., 25, 2011
|
|
2YHO
 
 | | The IDOL-UBE2D complex mediates sterol-dependent degradation of the LDL receptor | | Descriptor: | ACETATE ION, E3 UBIQUITIN-PROTEIN LIGASE MYLIP, UBIQUITIN-CONJUGATING ENZYME E2 D1, ... | | Authors: | Fairall, L, Goult, B.T, Millard, C.J, Tontonoz, P, Schwabe, J.W.R. | | Deposit date: | 2011-05-04 | | Release date: | 2011-06-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Idol-Ube2D Complex Mediates Sterol-Dependent Degradation of the Ldl Receptor.
Genes Dev., 25, 2011
|
|
7VQD
 
 | | Structure of MA1831 from Methanosarcina acetivorans in complex with farnesyl pyrophosphate and geranylgeranyl pyrophosphate. | | Descriptor: | Di-trans-poly-cis-decaprenylcistransferase, FARNESYL DIPHOSPHATE, NerylNeryl pyrophosphate, ... | | Authors: | Zhang, L.L, Chen, C.C, Liu, W.D, Huang, J.W, Zhang, X.W, Liu, B.B, Guo, R.T. | | Deposit date: | 2021-10-19 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structural insights to a bi-functional isoprenyl diphosphate synthase that can catalyze head-to-tail and head-to-middle condensation.
Int.J.Biol.Macromol., 214, 2022
|
|
7VQC
 
 | | Structure of MA1831 from Methanosarcina acetivorans in complex with pyrophosphate | | Descriptor: | Di-trans-poly-cis-decaprenylcistransferase, PYROPHOSPHATE 2-, SULFATE ION | | Authors: | Zhang, L.L, Chen, C.C, Liu, W.D, Huang, J.W, Zhang, X.W, Liu, B.B, Guo, R.T. | | Deposit date: | 2021-10-19 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural insights to a bi-functional isoprenyl diphosphate synthase that can catalyze head-to-tail and head-to-middle condensation.
Int.J.Biol.Macromol., 214, 2022
|
|
7VQB
 
 | | Structure of MA1831 from Methanosarcina acetivorans in complex with farnesyl pyrophosphate and dimethylallyl diphosphate | | Descriptor: | DIMETHYLALLYL DIPHOSPHATE, Di-trans-poly-cis-decaprenylcistransferase, FARNESYL DIPHOSPHATE, ... | | Authors: | Zhang, L.L, Chen, C.C, Liu, W.D, Huang, J.W, Zhang, X.W, Liu, B.B, Guo, R.T. | | Deposit date: | 2021-10-19 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structural insights to a bi-functional isoprenyl diphosphate synthase that can catalyze head-to-tail and head-to-middle condensation.
Int.J.Biol.Macromol., 214, 2022
|
|
7VQA
 
 | | Structure of MA1831 from Methanosarcina acetivorans in complex with dimethylallyl diphosphate. | | Descriptor: | DIMETHYLALLYL DIPHOSPHATE, Di-trans-poly-cis-decaprenylcistransferase, MAGNESIUM ION, ... | | Authors: | Zhang, L.L, Chen, C.C, Liu, W.D, Huang, J.W, Zhang, X.W, Liu, B.B, Guo, R.T. | | Deposit date: | 2021-10-19 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural insights to a bi-functional isoprenyl diphosphate synthase that can catalyze head-to-tail and head-to-middle condensation.
Int.J.Biol.Macromol., 214, 2022
|
|
7VQ9
 
 | | Structure of MA1831 from Methanosarcina acetivorans in complex with farnesyl thiopyrophosphate and isopentyl S-thiolodiphosphate | | Descriptor: | 3-methylbut-3-enylsulfanyl(phosphonooxy)phosphinic acid, Di-trans-poly-cis-decaprenylcistransferase, MAGNESIUM ION, ... | | Authors: | Zhang, L.L, Chen, C.C, Liu, W.D, Huang, J.W, Zhang, X.W, Liu, B.B, Guo, R.T. | | Deposit date: | 2021-10-19 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural insights to a bi-functional isoprenyl diphosphate synthase that can catalyze head-to-tail and head-to-middle condensation.
Int.J.Biol.Macromol., 214, 2022
|
|
6O3K
 
 | |
6O3U
 
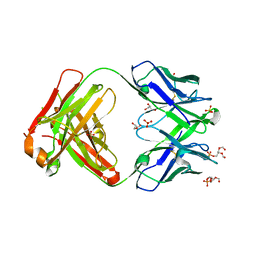 | |
6O3D
 
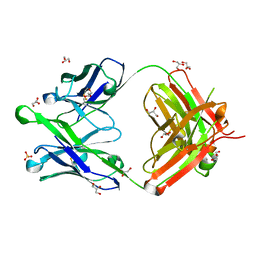 | |
6O3J
 
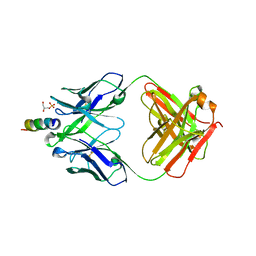 | | Crystal structure of the Fab fragment of the human HIV-1 neutralizing antibody PGZL1 in complex with its MPER peptide epitope (region 671-683 of HIV-1 gp41) and phosphatidic acid (06:0 PA) | | Descriptor: | (2R)-3-(phosphonooxy)propane-1,2-diyl dihexanoate, (4S)-2-METHYL-2,4-PENTANEDIOL, MPER peptide, ... | | Authors: | Irimia, A, Wilson, I.A. | | Deposit date: | 2019-02-26 | | Release date: | 2019-12-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.416 Å) | | Cite: | An MPER antibody neutralizes HIV-1 using germline features shared among donors.
Nat Commun, 10, 2019
|
|
6O3L
 
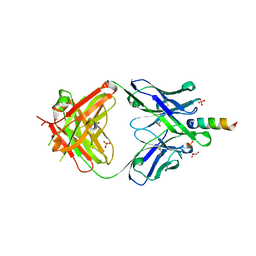 | |
6O41
 
 | |
6O42
 
 | |
3WUY
 
 | | Crystal structure of Nit6803 | | Descriptor: | Nitrilase | | Authors: | Yuan, Y.A, Yin, B, Wang, C. | | Deposit date: | 2014-05-09 | | Release date: | 2014-12-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural insights into enzymatic activity and substrate specificity determination by a single amino acid in nitrilase from Syechocystis sp. PCC6803
J.Struct.Biol., 188, 2014
|
|
8KI5
 
 | | PhmA, a type I diterpene synthase without NST/DTE motif | | Descriptor: | (2Z,6Z)-3,7,11-trimethyldodeca-2,6,10-trien-1-ol, PhmA | | Authors: | Zhang, B, Ge, H.M, Zhu, A, Zhang, Y. | | Deposit date: | 2023-08-22 | | Release date: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Biosynthesis of Platelet Activating Factor Antagonist Phomactins Revealing a New Class of Type I Diterpene Synthase
To Be Published
|
|
