7OPB
 
 | | IL7R in complex with an antagonist | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, IL7R binder, ... | | Authors: | Markovic, I, Verschueren, K.H.G, Verstraete, K, Savvides, S.N. | | Deposit date: | 2021-05-31 | | Release date: | 2022-05-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.144 Å) | | Cite: | Design of protein-binding proteins from the target structure alone.
Nature, 605, 2022
|
|
2SXL
 
 | | SEX-LETHAL RBD1, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | SEX-LETHAL PROTEIN | | Authors: | Inoue, M, Muto, Y, Sakamoto, H, Kigawa, T, Takio, K, Shimura, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1997-07-16 | | Release date: | 1998-07-22 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A characteristic arrangement of aromatic amino acid residues in the solution structure of the amino-terminal RNA-binding domain of Drosophila sex-lethal.
J.Mol.Biol., 272, 1997
|
|
1SXL
 
 | |
2GQG
 
 | | X-ray Crystal Structure of Dasatinib (BMS-354825) Bound to Activated ABL Kinase Domain | | Descriptor: | GLYCEROL, N-(2-CHLORO-6-METHYLPHENYL)-2-({6-[4-(2-HYDROXYETHYL)PIPERAZIN-1-YL]-2-METHYLPYRIMIDIN-4-YL}AMINO)-1,3-THIAZOLE-5-CARBOXAMIDE, Proto-oncogene tyrosine-protein kinase ABL1 | | Authors: | Klei, H.E. | | Deposit date: | 2006-04-20 | | Release date: | 2006-11-21 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Structure of Dasatinib (BMS-354825) Bound to Activated ABL Kinase Domain Elucidates Its Inhibitory Activity against Imatinib-Resistant ABL Mutants
CANCER RES., 66, 2006
|
|
7UDK
 
 | | Crystal structure of designed helical repeat protein RPB_LRP2_R4 bound to LRPx4 peptide | | Descriptor: | 4xLRP, Designed helical repeat protein (DHR) RPB_LRP2_R4 | | Authors: | Chang, Y, Redler, R.L, Bhabha, G, Ekiert, D.C. | | Deposit date: | 2022-03-20 | | Release date: | 2023-03-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | De novo design of modular peptide-binding proteins by superhelical matching.
Nature, 616, 2023
|
|
7UDL
 
 | | Crystal structure of designed helical repeat protein RPB_PLP1_R6 bound to PLPx6 peptide | | Descriptor: | 1,2-ETHANEDIOL, 6xPLP Peptide, Designed helical repeat protein (DHR) RPB_PLP1_R6 | | Authors: | Chang, Y, Redler, R.L, Bhabha, G, Ekiert, D.C. | | Deposit date: | 2022-03-20 | | Release date: | 2023-03-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | De novo design of modular peptide-binding proteins by superhelical matching.
Nature, 616, 2023
|
|
7UDN
 
 | |
7N1J
 
 | |
7N1K
 
 | |
7N3T
 
 | | TrkA ECD complex with designed miniprotein ligand | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Jude, K.M, Cao, L, Garcia, K.C. | | Deposit date: | 2021-06-01 | | Release date: | 2022-04-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Design of protein-binding proteins from the target structure alone.
Nature, 605, 2022
|
|
7UDM
 
 | | Crystal structure of designed helical repeat protein RPB_PLP1_R6 in alternative conformation 1 (with peptide) | | Descriptor: | 6xPLP, Designed helical repeat protein (DHR) RPB_PLP1_R6 | | Authors: | Chang, Y, Redler, R.L, Bhabha, G, Ekiert, D.C. | | Deposit date: | 2022-03-20 | | Release date: | 2023-03-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | De novo design of modular peptide-binding proteins by superhelical matching.
Nature, 616, 2023
|
|
7UDO
 
 | | Crystal structure of designed helical repeat protein RPB_LRP2_R4 (proteolysis fragment?), forming pseudopolymeric filaments | | Descriptor: | 1,2-ETHANEDIOL, Designed helical repeat protein (DHR) RPB_LRP2_R4, PHOSPHATE ION | | Authors: | Redler, R.L, Chang, Y, Bhabha, G, Ekiert, D.C. | | Deposit date: | 2022-03-20 | | Release date: | 2023-03-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | De novo design of modular peptide-binding proteins by superhelical matching.
Nature, 616, 2023
|
|
7UDJ
 
 | | Crystal structure of designed helical repeat protein RPB_PEW3_R4 bound to PAWx4 peptide | | Descriptor: | 4xPAW peptide, De novo designed helical repeat protein RPB_PEW3_R4 | | Authors: | Redler, R.L, Chang, Y, Bhabha, G, Ekiert, D. | | Deposit date: | 2022-03-20 | | Release date: | 2023-03-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | De novo design of modular peptide-binding proteins by superhelical matching.
Nature, 616, 2023
|
|
7UE2
 
 | |
2OJ9
 
 | | Structure of IGF-1R kinase domain complexed with a benzimidazole inhibitor | | Descriptor: | 3-[5-(1H-IMIDAZOL-1-YL)-7-METHYL-1H-BENZIMIDAZOL-2-YL]-4-[(PYRIDIN-2-YLMETHYL)AMINO]PYRIDIN-2(1H)-ONE, Insulin-like growth factor 1 receptor precursor (EC 2.7.10.1) (Insulin-like growth factor I receptor) (IGF-I receptor) (CD221 antigen) | | Authors: | Sack, J.S, Jacobson, B.L. | | Deposit date: | 2007-01-12 | | Release date: | 2007-05-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery and initial SAR of 3-(1H-benzo[d]imidazol-2-yl)pyridin-2(1H)-ones as inhibitors of insulin-like growth factor 1-receptor (IGF-1R).
Bioorg.Med.Chem.Lett., 17, 2007
|
|
4LRG
 
 | | Structure of BRD4 bromodomain 1 with a dimethyl thiophene isoxazole azepine carboxamide | | Descriptor: | 2-[(6S)-4-(4-chlorophenyl)-2,3,9-trimethyl-6H-[1,2]oxazolo[5,4-c]thieno[2,3-e]azepin-6-yl]acetamide, Bromodomain-containing protein 4 | | Authors: | Ravichandran, S, Jayaram, H, Poy, F, Gehling, V, Hewitt, M, Vaswani, R, Leblanc, Y, Cote, A, Nasveschuk, C, Taylor, A, Harmange, J.-C, Audia, J, Pardo, E, Joshi, S, Sandy, P, Mertz, J, Sims, R, Bergeron, L, Bryant, B, Yellapuntala, S, Nandana, B.S, Birudukota, S, Albrecht, B, Bellon, S. | | Deposit date: | 2013-07-19 | | Release date: | 2013-08-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Discovery, Design, and Optimization of Isoxazole Azepine BET Inhibitors.
ACS Med Chem Lett, 4, 2013
|
|
4LR6
 
 | | Structure of BRD4 bromodomain 1 with a 3-methyl-4-phenylisoxazol-5-amine fragment | | Descriptor: | 3-methyl-4-phenyl-1,2-oxazol-5-amine, Bromodomain-containing protein 4, FORMIC ACID | | Authors: | Jayaram, H, Poy, F, Gehling, V, Hewitt, M, Vaswani, R, Leblanc, Y, Cote, A, Nasveschuk, C, Taylor, A, Harmange, J.-C, Audia, J, Pardo, E, Joshi, S, Sandy, P, Mertz, J, Sims, R, Bergeron, L, Bryant, B, Ravichandran, S, Yellapuntala, S, Nandana, B.S, Birudukota, S, Albrecht, B, Bellon, S. | | Deposit date: | 2013-07-19 | | Release date: | 2013-08-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Discovery, Design, and Optimization of Isoxazole Azepine BET Inhibitors.
ACS Med Chem Lett, 4, 2013
|
|
7RDH
 
 | |
7RKC
 
 | | Computationally designed tunable C2 symmetric tandem repeat homodimer, D_3_633 | | Descriptor: | ACETATE ION, D_3_633 | | Authors: | Kennedy, M.A, Stoddard, B.L, Hicks, D.R, Bera, A.K. | | Deposit date: | 2021-07-22 | | Release date: | 2022-05-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | De novo design of protein homodimers containing tunable symmetric protein pockets.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7RMY
 
 | | De Novo designed tunable protein pockets, D_3-337 | | Descriptor: | De Novo designed tunable homodimer, D_3-337 | | Authors: | Bera, A.K, Hicks, D.R, Kang, A, Sankaran, B, Baker, D. | | Deposit date: | 2021-07-28 | | Release date: | 2022-08-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | De novo design of protein homodimers containing tunable symmetric protein pockets.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7RMX
 
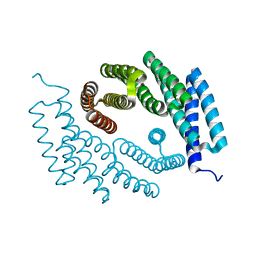 | | Structure of De Novo designed tunable symmetric protein pockets | | Descriptor: | Tunable symmetric protein, D_3_212 | | Authors: | Bera, A.K, Hicks, D.R, Kang, A, Sankaran, B, Baker, D. | | Deposit date: | 2021-07-28 | | Release date: | 2022-08-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | De novo design of protein homodimers containing tunable symmetric protein pockets.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6AR4
 
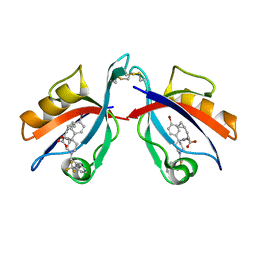 | | Crystal structure of PICK1 in complex with the small molecule inhibitor 1o | | Descriptor: | N-[4-(4-bromophenyl)-1-{[2-(trifluoromethyl)phenyl]methyl}piperidine-4-carbonyl]-3-cyclopropyl-L-alanine, PRKCA-binding protein | | Authors: | Marcotte, D. | | Deposit date: | 2017-08-21 | | Release date: | 2018-08-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Potent PDZ-Domain PICK1 Inhibitors that Modulate Amyloid Beta-Mediated Synaptic Dysfunction.
Sci Rep, 8, 2018
|
|
7S5B
 
 | |
2FXD
 
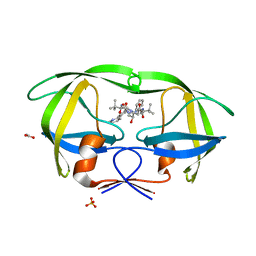 | | X-ray crystal structure of HIV-1 protease IRM mutant complexed with atazanavir (BMS-232632) | | Descriptor: | (3S,8S,9S,12S)-3,12-BIS(1,1-DIMETHYLETHYL)-8-HYDROXY-4,11-DIOXO-9-(PHENYLMETHYL)-6-[[4-(2-PYRIDINYL)PHENYL]METHYL]-2,5, 6,10,13-PENTAAZATETRADECANEDIOIC ACID DIMETHYL ESTER, ACETATE ION, ... | | Authors: | Klei, H.E, Sheriff, S. | | Deposit date: | 2006-02-04 | | Release date: | 2007-02-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray crystal structures of human immunodeficiency virus type 1 protease mutants complexed with atazanavir.
J.Virol., 81, 2007
|
|
2FXE
 
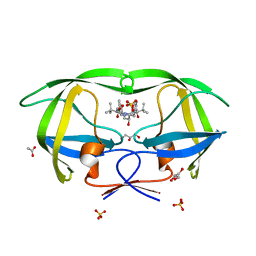 | | X-ray crystal structure of HIV-1 protease CRM mutant complexed with atazanavir (BMS-232632) | | Descriptor: | (3S,8S,9S,12S)-3,12-BIS(1,1-DIMETHYLETHYL)-8-HYDROXY-4,11-DIOXO-9-(PHENYLMETHYL)-6-[[4-(2-PYRIDINYL)PHENYL]METHYL]-2,5, 6,10,13-PENTAAZATETRADECANEDIOIC ACID DIMETHYL ESTER, ACETATE ION, ... | | Authors: | Sheriff, S, Klei, H.E. | | Deposit date: | 2006-02-05 | | Release date: | 2007-02-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystal structures of human immunodeficiency virus type 1 protease mutants complexed with atazanavir.
J.Virol., 81, 2007
|
|
