8CKC
 
 | | Vitamin D receptor complex with 25-amine derivative of 1,25D3 | | Descriptor: | (1R,3S,5Z)-5-[(2E)-2-[(1R,3aS,7aR)-1-[(2R)-6-azanyl-6-methyl-heptan-2-yl]-7a-methyl-2,3,3a,5,6,7-hexahydro-1H-inden-4-ylidene]ethylidene]-4-methylidene-cyclohexane-1,3-diol, ACETATE ION, Nuclear receptor coactivator 1, ... | | Authors: | Rochel, N. | | Deposit date: | 2023-02-15 | | Release date: | 2023-04-26 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural analysis and biological activities of C25-amino and C25-nitro vitamin D analogs.
Bioorg.Chem., 136, 2023
|
|
8CK5
 
 | | VDR LBD complex with 25-nitro derivative of 1,25D3 | | Descriptor: | (1R,3S,5Z)-5-[(2E)-2-[(1R,3aS,7aR)-7a-methyl-1-[(2R)-6-methyl-6-nitro-heptan-2-yl]-2,3,3a,5,6,7-hexahydro-1H-inden-4-ylidene]ethylidene]-4-methylidene-cyclohexane-1,3-diol, ACETATE ION, Nuclear receptor coactivator 1, ... | | Authors: | Rochel, N. | | Deposit date: | 2023-02-14 | | Release date: | 2023-04-26 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural analysis and biological activities of C25-amino and C25-nitro vitamin D analogs.
Bioorg.Chem., 136, 2023
|
|
8R3D
 
 | | Crystal structure of Pent only at pH 8.8 | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, Beta propeller | | Authors: | Flood, R.J, Crowley, P.B. | | Deposit date: | 2023-11-08 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Multivalent Calixarene Complexation of a Designed Pentameric Lectin.
Biomacromolecules, 25, 2024
|
|
4CCU
 
 | | Structure of the Human Anaplastic Lymphoma Kinase in Complex with 2-(5-(6-amino-5-((R)-1-(5-fluoro-2-(2H-1,2,3-triazol-2-yl)phenyl)ethoxy) pyridin-3-yl)-4-methylthiazol-2-yl)propan-2-ol | | Descriptor: | 2-(5-(6-amino-5-((R)-1-(5-fluoro-2-(2H-1,2,3-triazol-2-yl)phenyl)ethoxy)pyridin-3-yl)-4-methylthiazol-2-yl)propan-2-ol, ALK TYROSINE KINASE RECEPTOR | | Authors: | McTigue, M, Deng, Y, Liu, W, Brooun, A, Stewart, A. | | Deposit date: | 2013-10-28 | | Release date: | 2014-01-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Design of Potent and Selective Inhibitors to Overcome Clinical Alk Mutations Resistant to Crizotinib.
J.Med.Chem., 57, 2014
|
|
8R3B
 
 | |
3KV0
 
 | | Crystal structure of HET-C2: A FUNGAL GLYCOLIPID TRANSFER PROTEIN (GLTP) | | Descriptor: | HET-C2 | | Authors: | Simanshu, D.K, Kenoth, R, Brown, R.E, Patel, D.J. | | Deposit date: | 2009-11-28 | | Release date: | 2010-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural determination and tryptophan fluorescence of heterokaryon incompatibility C2 protein (HET-C2), a fungal glycolipid transfer protein (GLTP), provide novel insights into glycolipid specificity and membrane interaction by the GLTP fold.
J.Biol.Chem., 285, 2010
|
|
4CNH
 
 | | Structure of the Human Anaplastic Lymphoma Kinase in Complex with the inhibitor 3-((1R)-1-(5-fluoro-2-methoxyphenyl)ethoxy)-5-(1-methyl-1H- 1,2,3-triazol-5-yl)pyridin-2-amine | | Descriptor: | 3-[(1R)-1-(5-fluoro-2-methoxyphenyl)ethoxy]-5-(1-methyl-1H-1,2,3-triazol-5-yl)pyridin-2-amine, ALK TYROSINE KINASE RECEPTOR | | Authors: | McTigue, M.A, Deng, Y.L, Liu, W, Brooun, A, Stewart, A.E. | | Deposit date: | 2014-01-22 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of (10R)-7-Amino-12-Fluoro-2,10,16-Trimethyl-15-Oxo-10,15,16,17-Tetrahydro-2H-8,4-(Metheno)Pyrazolo[4,3-H][2,5,11]Benzoxadiazacyclotetradecine-3-Carbonitrile (Pf-06463922), a Macrocyclic Inhibitor of Alk/Ros1 with Pre-Clinical Brain Exposure and Broad Spectrum Potency Against Alk-Resistant Mutations.
J.Med.Chem., 57, 2014
|
|
4CLI
 
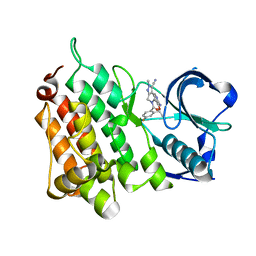 | | Structure of the Human Anaplastic Lymphoma Kinase in Complex with PF- 06463922 ((10R)-7-amino-12-fluoro-2,10,16-trimethyl-15-oxo-10,15,16, 17-tetrahydro-2H-8,4-(metheno)pyrazolo(4,3-h)(2,5,11) benzoxadiazacyclotetradecine-3-carbonitrile). | | Descriptor: | (10R)-7-amino-12-fluoro-2,10,16-trimethyl-15-oxo-10,15,16,17-tetrahydro-2H-8,4-(metheno)pyrazolo[4,3-h][2,5,11]benzoxadiazacyclotetradecine-3-carbonitrile, ALK TYROSINE KINASE RECEPTOR | | Authors: | McTigue, M.A, Deng, Y.L, Liu, W, Brooun, A, Stewart, A.E. | | Deposit date: | 2014-01-14 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery of (10R)-7-Amino-12-Fluoro-2,10,16-Trimethyl-15-Oxo-10,15,16,17-Tetrahydro-2H-8,4-(Metheno)Pyrazolo[4,3-H][2,5,11]Benzoxadiazacyclotetradecine-3-Carbonitrile (Pf-06463922), a Macrocyclic Inhibitor of Alk/Ros1 with Pre-Clinical Brain Exposure and Broad Spectrum Potency Against Alk-Resistant Mutations.
J.Med.Chem., 57, 2014
|
|
4CMT
 
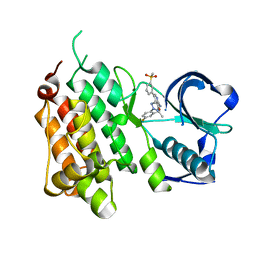 | | Structure of the Human Anaplastic Lymphoma Kinase in Complex with the inhibitor 3-((1R)-1-(5-fluoro-2-(2H-1,2,3-triazol-2-yl)phenyl)ethoxy)- 5-(3-(methylsulfonyl)phenyl)pyridin-2-amine | | Descriptor: | 3-{(1R)-1-[2-(1,3-dihydro-2H-1,2,3-triazol-2-yl)-5-fluorophenyl]ethoxy}-5-[3-(methylsulfonyl)phenyl]pyridin-2-amine, ALK TYROSINE KINASE RECEPTOR | | Authors: | McTigue, M.A, Deng, Y.L, Liu, W, Brooun, A, Stewart, A.E. | | Deposit date: | 2014-01-17 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Discovery of (10R)-7-Amino-12-Fluoro-2,10,16-Trimethyl-15-Oxo-10,15,16,17-Tetrahydro-2H-8,4-(Metheno)Pyrazolo[4,3-H][2,5,11]Benzoxadiazacyclotetradecine-3-Carbonitrile (Pf-06463922), a Macrocyclic Inhibitor of Alk/Ros1 with Pre-Clinical Brain Exposure and Broad Spectrum Potency Against Alk-Resistant Mutations.
J.Med.Chem., 57, 2014
|
|
4CCB
 
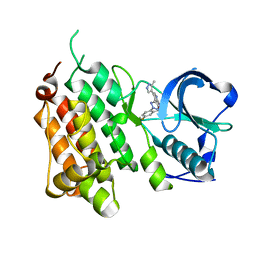 | | Structure of the Human Anaplastic Lymphoma Kinase in Complex with 3-((R)-1-(5-fluoro-2-(2H-1,2,3-triazol-2-yl)phenyl)ethoxy)-5-(5-methyl-1H- pyrazol-4-yl)pyridin-2-amine | | Descriptor: | 3-[(1R)-1-[5-fluoranyl-2-(1,2,3-triazol-2-yl)phenyl]ethoxy]-5-(3-methyl-1H-pyrazol-4-yl)pyridin-2-amine, ALK TYROSINE KINASE RECEPTOR | | Authors: | McTigue, M, Deng, Y, Liu, W, Brooun, A, Stewart, A. | | Deposit date: | 2013-10-21 | | Release date: | 2014-01-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | The Design of Potent and Selective Inhibitors to Overcome Clinical Alk Mutations Resistant to Crizotinib.
J.Med.Chem., 57, 2014
|
|
4CD0
 
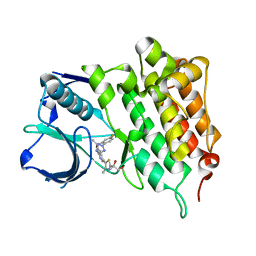 | | Structure of L1196M Mutant Human Anaplastic Lymphoma Kinase in Complex with 2-(5-(6-amino-5-((R)-1-(5-fluoro-2-(2H-1,2,3-triazol-2- yl)phenyl)ethoxy)pyridin-3-yl)-4-methylthiazol-2-yl)propane-1,2-diol | | Descriptor: | (2R)-2-[5-(6-amino-5-{(1R)-1-[2-(1,3-dihydro-2H-1,2,3-triazol-2-yl)-5-fluorophenyl]ethoxy}pyridin-3-yl)-4-methyl-1,3-thiazol-2-yl]propane-1,2-diol, ALK TYROSINE KINASE RECEPTOR | | Authors: | McTigue, M, Deng, Y, Liu, W, Brooun, A, Stewart, A. | | Deposit date: | 2013-10-29 | | Release date: | 2014-01-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | The Design of Potent and Selective Inhibitors to Overcome Clinical Alk Mutations Resistant to Crizotinib.
J.Med.Chem., 57, 2014
|
|
6TS6
 
 | | Coagulation factor XI protease domain in complex with active site inhibitor | | Descriptor: | 2-[2-[[3-[(3~{S})-3-azanyl-2,3-dihydro-1-benzofuran-5-yl]-5-(2-cyanopropan-2-yl)phenyl]methoxy]phenyl]ethanoic acid, Coagulation factor XI, DIMETHYL SULFOXIDE, ... | | Authors: | Renatus, M, Schiering, N. | | Deposit date: | 2019-12-20 | | Release date: | 2020-07-08 | | Last modified: | 2020-08-26 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Structure-Based Design and Preclinical Characterization of Selective and Orally Bioavailable Factor XIa Inhibitors: Demonstrating the Power of an Integrated S1 Protease Family Approach.
J.Med.Chem., 63, 2020
|
|
6TS4
 
 | | Coagulation factor XI protease domain in complex with active site inhibitor | | Descriptor: | 2-[2-[[3-[3-(aminomethyl)phenyl]phenyl]carbonylamino]phenyl]ethanoic acid, Coagulation factor XI, DIMETHYL SULFOXIDE, ... | | Authors: | Renatus, M, Schiering, N. | | Deposit date: | 2019-12-19 | | Release date: | 2020-07-08 | | Last modified: | 2020-08-26 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Structure-Based Design and Preclinical Characterization of Selective and Orally Bioavailable Factor XIa Inhibitors: Demonstrating the Power of an Integrated S1 Protease Family Approach.
J.Med.Chem., 63, 2020
|
|
6TS7
 
 | | Coagulation factor XI protease domain in complex with active site inhibitor | | Descriptor: | 2-[2-[[3-(1,2,3,4-tetrahydroisoquinolin-7-yl)phenyl]methoxy]phenyl]ethanoic acid, Coagulation factor XI | | Authors: | Renatus, M, Schiering, N. | | Deposit date: | 2019-12-20 | | Release date: | 2020-07-08 | | Last modified: | 2020-08-26 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Structure-Based Design and Preclinical Characterization of Selective and Orally Bioavailable Factor XIa Inhibitors: Demonstrating the Power of an Integrated S1 Protease Family Approach.
J.Med.Chem., 63, 2020
|
|
6USY
 
 | | COAGULATION FACTOR XI CATALYTIC DOMAIN (C123S) IN COMPLEX WITH NVP-XIV936 | | Descriptor: | 1-[(2S)-2-{3-[(3S)-3-amino-2,3-dihydro-1-benzofuran-5-yl]-5-(propan-2-yl)phenyl}-2-hydroxyethyl]-1H-indole-7-carboxylic acid, Coagulation factor XIa light chain | | Authors: | Weihofen, W.A, Clark, K, Nunes, S. | | Deposit date: | 2019-10-28 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Structure-Based Design and Preclinical Characterization of Selective and Orally Bioavailable Factor XIa Inhibitors: Demonstrating the Power of an Integrated S1 Protease Family Approach.
J.Med.Chem., 63, 2020
|
|
6XTV
 
 | | FULL-LENGTH LTTR LYSG FROM CORYNEBACTERIUM GLUTAMICUM WITH BOUND EFFECTOR ARG | | Descriptor: | ARGININE, Lysine export transcriptional regulatory protein LysG | | Authors: | Hofmann, E, Syberg, F, Schlicker, C, Eggeling, L, Schendzielorz, G. | | Deposit date: | 2020-01-16 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Engineering and application of a biosensor with focused ligand specificity.
Nat Commun, 11, 2020
|
|
6XTU
 
 | | FULL-LENGTH LTTR LYSG FROM CORYNEBACTERIUM GLUTAMICUM | | Descriptor: | Lysine export transcriptional regulatory protein LysG | | Authors: | Hofmann, E, Syberg, F, Schlicker, C, Eggeling, L, Schendzielorz, G. | | Deposit date: | 2020-01-16 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Engineering and application of a biosensor with focused ligand specificity.
Nat Commun, 11, 2020
|
|
6HQU
 
 | | Humanised RadA mutant HumRadA22 in complex with a recombined BRC repeat 8-2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Breast cancer type 2 susceptibility, DNA repair and recombination protein RadA, ... | | Authors: | Pantelejevs, T, Lindenburg, L, Hyvonen, M, Hollfelder, F. | | Deposit date: | 2018-09-25 | | Release date: | 2019-10-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Improved RAD51 binders through motif shuffling based on the modularity of BRC repeats.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
1UNL
 
 | | Structural mechanism for the inhibition of CD5-p25 from the roscovitine, aloisine and indirubin. | | Descriptor: | CYCLIN-DEPENDENT KINASE 5, CYCLIN-DEPENDENT KINASE 5 ACTIVATOR 1, R-ROSCOVITINE | | Authors: | Mapelli, M, Crovace, C, Massimiliano, L, Musacchio, A. | | Deposit date: | 2003-09-10 | | Release date: | 2004-11-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanism of Cdk5/P25 Binding by Cdk Inhibitors
J.Med.Chem., 48, 2005
|
|
1UNG
 
 | | Structural mechanism for the inhibition of CDK5-p25 by roscovitine, aloisine and indirubin. | | Descriptor: | 6-PHENYL[5H]PYRROLO[2,3-B]PYRAZINE, CELL DIVISION PROTEIN KINASE 5, CYCLIN-DEPENDENT KINASE 5 ACTIVATOR 1 | | Authors: | Mapelli, M, Crovace, C, Massimiliano, L, Musacchio, A. | | Deposit date: | 2003-09-10 | | Release date: | 2004-11-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanism of Cdk5/P25 Binding by Cdk Inhibitors
J.Med.Chem., 48, 2005
|
|
1UNH
 
 | | Structural mechanism for the inhibition of CDK5-p25 by roscovitine, aloisine and indirubin. | | Descriptor: | (Z)-1H,1'H-[2,3']BIINDOLYLIDENE-3,2'-DIONE-3-OXIME, CYCLIN-DEPENDENT KINASE 5, CYCLIN-DEPENDENT KINASE 5 ACTIVATOR 1 | | Authors: | Mapelli, M, Crovace, C, Massimiliano, L, Musacchio, A. | | Deposit date: | 2003-09-10 | | Release date: | 2004-11-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Mechanism of Cdk5/P25 Binding by Cdk Inhibitors
J.Med.Chem., 48, 2005
|
|
6UKM
 
 | | STING C-terminal Domain Complexed with Non-cyclic Dinucleotide Compound MSA-2 | | Descriptor: | 4-(5,6-dimethoxy-1-benzothiophen-2-yl)-4-oxobutanoic acid, fusion protein of Ubiquitin-like protein SMT3 and Stimulator of interferon protein c-terminal domain | | Authors: | Lesburg, C.A. | | Deposit date: | 2019-10-05 | | Release date: | 2020-08-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | An orally available non-nucleotide STING agonist with antitumor activity.
Science, 369, 2020
|
|
6UKU
 
 | | STING C-terminal Domain Complexed with Non-cyclic Dinucleotide Compound 3 | | Descriptor: | 4,4'-[propane-1,3-diylbis(6-methoxy-1-benzothiene-5,2-diyl)]bis(4-oxobutanoic acid), fusion protein of Ubiquitin-like protein SMT3 and Stimulator of interferon protein c-terminal domain | | Authors: | Lesburg, C.A. | | Deposit date: | 2019-10-06 | | Release date: | 2020-08-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | An orally available non-nucleotide STING agonist with antitumor activity.
Science, 369, 2020
|
|
6UL0
 
 | | STING C-terminal Domain Complexed with Non-cyclic Dinucleotide Compound 4 | | Descriptor: | 4-{5-[(1Z)-3-{[2-(3-carboxypropanoyl)-6-methoxy-1-benzothiophen-5-yl]oxy}prop-1-en-1-yl]-6-methoxy-1-benzothiophen-2-yl}-4-oxobutanoic acid, fusion protein of Ubiquitin-like protein SMT3 and Stimulator of interferon protein c-terminal domain | | Authors: | Lesburg, C.A. | | Deposit date: | 2019-10-06 | | Release date: | 2020-08-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | An orally available non-nucleotide STING agonist with antitumor activity.
Science, 369, 2020
|
|
5EXB
 
 | |
