4PD1
 
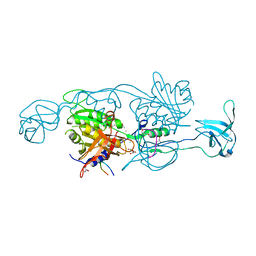 | | Structure of gephyrin E domain with Glycine-beta receptor peptide | | Descriptor: | ACETATE ION, GLYCEROL, Gephyrin, ... | | Authors: | Kasaragod, V.B, Maric, H.M, Schindelin, H. | | Deposit date: | 2014-04-17 | | Release date: | 2014-08-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.975 Å) | | Cite: | Modulation of gephyrin-glycine receptor affinity by multivalency.
Acs Chem.Biol., 9, 2014
|
|
7E5F
 
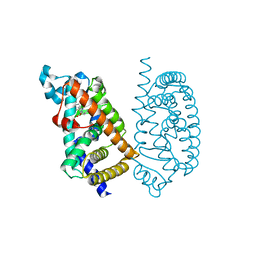 | | HUMAN PPAR ALPHA LIGAND BINDING DOMAIN IN COMPLEX WITH TIPP703 OBTAINED BY SOAKING | | Descriptor: | (2S)-2-(4-propoxy-3-{[({4-[(3S,5S,7S)-tricyclo[3.3.1.1~3,7~]dec-1-yl]phenyl}carbonyl)amino]methyl}benzyl)butanoic acid, Peroxisome proliferator-activated receptor alpha | | Authors: | Oyama, T, Kamata, S, Ishii, I, Miyachi, H. | | Deposit date: | 2021-02-18 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal Structures of the Human Peroxisome Proliferator-Activated Receptor (PPAR) alpha Ligand-Binding Domain in Complexes with a Series of Phenylpropanoic Acid Derivatives Generated by a Ligand-Exchange Soaking Method.
Biol.Pharm.Bull., 44, 2021
|
|
7E5G
 
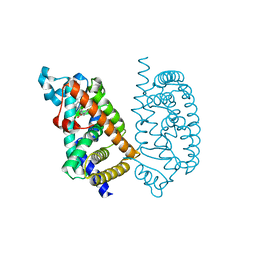 | | HUMAN PPAR ALPHA LIGAND BINDING DOMAIN IN COMPLEX WITH YN4pai OBTAINED BY SOAKING | | Descriptor: | (2S)-2-[[4-butoxy-3-[(pyren-1-ylcarbonylamino)methyl]phenyl]methyl]butanoic acid, Peroxisome proliferator-activated receptor alpha | | Authors: | Oyama, T, Kamata, S, Ishii, I, Miyachi, H. | | Deposit date: | 2021-02-18 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Crystal Structures of the Human Peroxisome Proliferator-Activated Receptor (PPAR) alpha Ligand-Binding Domain in Complexes with a Series of Phenylpropanoic Acid Derivatives Generated by a Ligand-Exchange Soaking Method.
Biol.Pharm.Bull., 44, 2021
|
|
7E5I
 
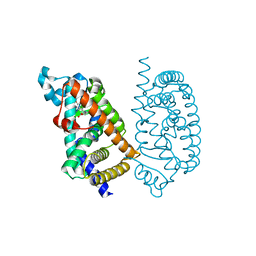 | | HUMAN PPAR ALPHA LIGAND BINDING DOMAIN IN COMPLEX WITH APHM6 OBTAINED BY SOAKING | | Descriptor: | (2S)-2-[[3-[[3-fluoranyl-4-(4-fluoranylphenoxy)phenyl]methylcarbamoyl]-4-methoxy-phenyl]methyl]butanoic acid, Peroxisome proliferator-activated receptor alpha | | Authors: | Oyama, T, Kamata, S, Ishii, I, Miyachi, H. | | Deposit date: | 2021-02-18 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal Structures of the Human Peroxisome Proliferator-Activated Receptor (PPAR) alpha Ligand-Binding Domain in Complexes with a Series of Phenylpropanoic Acid Derivatives Generated by a Ligand-Exchange Soaking Method.
Biol.Pharm.Bull., 44, 2021
|
|
7E5H
 
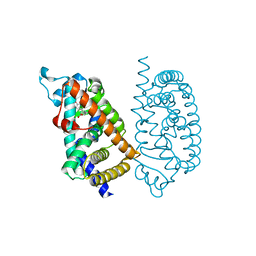 | | HUMAN PPAR ALPHA LIGAND BINDING DOMAIN IN COMPLEX WITH APHM6 OBTAINED BY COCRYSTALLIZATION | | Descriptor: | (2S)-2-[[3-[[3-fluoranyl-4-(4-fluoranylphenoxy)phenyl]methylcarbamoyl]-4-methoxy-phenyl]methyl]butanoic acid, Peroxisome proliferator-activated receptor alpha | | Authors: | Oyama, T, Kamata, S, Ishii, I, Miyachi, H. | | Deposit date: | 2021-02-18 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Crystal Structures of the Human Peroxisome Proliferator-Activated Receptor (PPAR) alpha Ligand-Binding Domain in Complexes with a Series of Phenylpropanoic Acid Derivatives Generated by a Ligand-Exchange Soaking Method.
Biol.Pharm.Bull., 44, 2021
|
|
5YL3
 
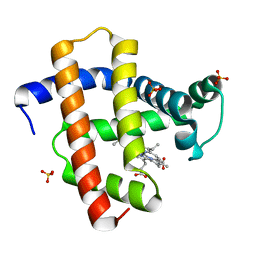 | | Crystal structure of horse heart myoglobin reconstituted with manganese porphycene in resting state at pH 8.5 | | Descriptor: | Myoglobin, PORPHYCENE CONTAINING MN, SULFATE ION | | Authors: | Oohora, K, Meichin, H, Kihira, Y, Sugimoto, H, Shiro, Y, Hayashi, T. | | Deposit date: | 2017-10-17 | | Release date: | 2017-12-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Manganese(V) Porphycene Complex Responsible for Inert C-H Bond Hydroxylation in a Myoglobin Matrix.
J. Am. Chem. Soc., 139, 2017
|
|
7EGO
 
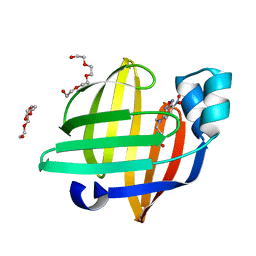 | | X-ray structure of the human heart fatty acid-binding protein complexed with the fluorescent probe HA527 | | Descriptor: | 3-[methyl-(4-nitro-2,1,3-benzoxadiazol-7-yl)amino]propanoic acid, Fatty acid-binding protein, heart, ... | | Authors: | Takabayashi, M, Yokota, J, Matsuoka, S, Tsuchikawa, H, Sonoyama, M, Inoue, Y, Hayashi, F, Sugiyama, S. | | Deposit date: | 2021-03-24 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | X-ray structure of the human heart fatty acid-binding protein complexed with the fluorescent probe HA527
To Be Published
|
|
3CMM
 
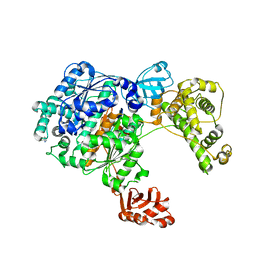 | | Crystal Structure of the Uba1-Ubiquitin Complex | | Descriptor: | PROLINE, Ubiquitin, Ubiquitin-activating enzyme E1 1 | | Authors: | Lee, I, Schindelin, H. | | Deposit date: | 2008-03-23 | | Release date: | 2008-08-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural insights into E1-catalyzed ubiquitin activation and transfer to conjugating enzymes.
Cell(Cambridge,Mass.), 134, 2008
|
|
1VCK
 
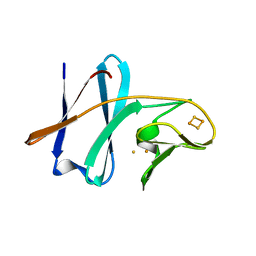 | | Crystal structure of ferredoxin component of carbazole 1,9a-dioxygenase of Pseudomonas resinovorans strain CA10 | | Descriptor: | FE (III) ION, FE2/S2 (INORGANIC) CLUSTER, HYDROSULFURIC ACID, ... | | Authors: | Nam, J.-W, Noguchi, H, Fujiomoto, Z, Mizuno, H, Fushinobu, S, Kobashi, N, Iwata, K, Yoshida, T, Habe, H, Yamane, H, Omori, T, Nojiri, H. | | Deposit date: | 2004-03-09 | | Release date: | 2005-03-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the ferredoxin component of carbazole 1,9a-dioxygenase of Pseudomonas resinovorans strain CA10, a novel Rieske non-heme iron oxygenase system
PROTEINS, 58, 2005
|
|
4F9Z
 
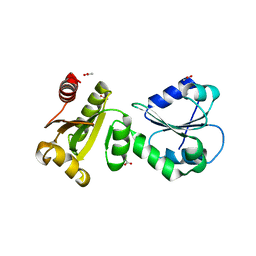 | | Crystal Structure of human ERp27 | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, ACETATE ION, ... | | Authors: | Kober, F.X, Koelmel, W, Kuper, J, Schindelin, H. | | Deposit date: | 2012-05-21 | | Release date: | 2012-12-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Crystal Structure of the Protein-Disulfide Isomerase Family Member ERp27 Provides Insights into Its Substrate Binding Capabilities.
J.Biol.Chem., 288, 2013
|
|
3M62
 
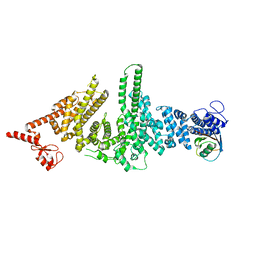 | |
6HRS
 
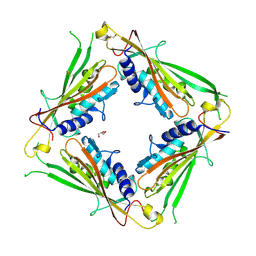 | | Structure of the TRPML2 ELD at pH 4.5 | | Descriptor: | GLYCEROL, Mucolipin-2 | | Authors: | Bader, N, Viet, K.K, Wagner, A, Hellmich, U.A, Schindelin, H. | | Deposit date: | 2018-09-28 | | Release date: | 2019-06-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure of the Human TRPML2 Ion Channel Extracytosolic/Lumenal Domain.
Structure, 27, 2019
|
|
5X7P
 
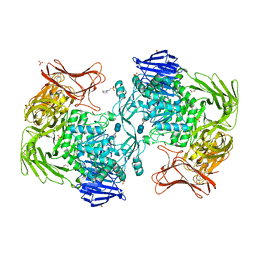 | | Crystal structure of Paenibacillus sp. 598K alpha-1,6-glucosyltransferase complexed with acarbose | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Fujimoto, Z, Kishine, N, Suzuki, N, Momma, M, Ichinose, H, Kimura, A, Funane, K. | | Deposit date: | 2017-02-27 | | Release date: | 2017-07-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Carbohydrate-binding architecture of the multi-modular alpha-1,6-glucosyltransferase from Paenibacillus sp. 598K, which produces alpha-1,6-glucosyl-alpha-glucosaccharides from starch
Biochem. J., 474, 2017
|
|
6ZQH
 
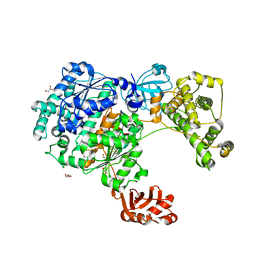 | | Yeast Uba1 in complex with ubiquitin | | Descriptor: | BETA-MERCAPTOETHANOL, GLYCEROL, TETRAETHYLENE GLYCOL, ... | | Authors: | Misra, M, Schindelin, H. | | Deposit date: | 2020-07-09 | | Release date: | 2020-11-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.032 Å) | | Cite: | Development of ADPribosyl Ubiquitin Analogues to Study Enzymes Involved in Legionella Infection.
Chemistry, 27, 2021
|
|
3D25
 
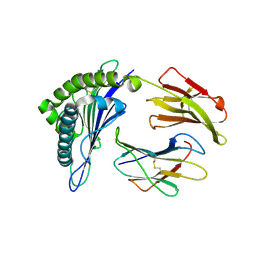 | | Crystal structure of HA-1 minor histocompatibility antigen bound to human class I MHC HLA-A2 | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-2 alpha chain, ... | | Authors: | Nicholls, S, Piper, K.P, Mohammed, F, Dafforn, T.R, Tenzer, S, Salim, M, Mahendra, P, Craddock, C, van Endert, P, Schild, H, Cobbold, M, Engelhard, V.H, Moss, P.A.H, Willcox, B.E. | | Deposit date: | 2008-05-07 | | Release date: | 2009-02-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Secondary anchor polymorphism in the HA-1 minor histocompatibility antigen critically affects MHC stability and TCR recognition
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
5X7R
 
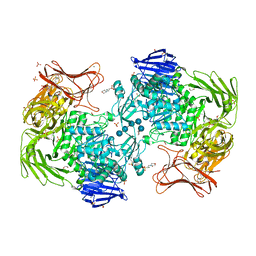 | | Crystal structure of Paenibacillus sp. 598K alpha-1,6-glucosyltransferase complexed with isomaltohexaose | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Fujimoto, Z, Kishine, N, Suzuki, N, Momma, M, Ichinose, H, Kimura, A, Funane, K. | | Deposit date: | 2017-02-27 | | Release date: | 2017-07-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Carbohydrate-binding architecture of the multi-modular alpha-1,6-glucosyltransferase from Paenibacillus sp. 598K, which produces alpha-1,6-glucosyl-alpha-glucosaccharides from starch
Biochem. J., 474, 2017
|
|
7FDX
 
 | | X-ray structure of the human heart fatty acid-binding protein complexed with the fluorescent probe 8-Anilino-1-naphthalenesulfonic acid (ANS) | | Descriptor: | 8-ANILINO-1-NAPHTHALENE SULFONATE, Fatty acid-binding protein, heart | | Authors: | Sugiyama, S, Matsuoka, S, Tsuchikawa, H, Sonoyama, M, Inoue, Y, Hayashi, F, Murata, M. | | Deposit date: | 2021-07-18 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | X-ray structure of the human heart fatty acid-binding protein complexed with the fluorescent probe 8-Anilino-1-naphthalenesulfonic acid (ANS)
To Be Published
|
|
7FFX
 
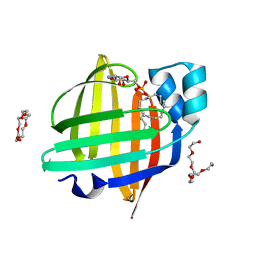 | | The 0.88 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with alpha-llinolenic acid | | Descriptor: | 2-amino-2-deoxy-6-O-phosphono-alpha-D-glucopyranose, ALPHA-LINOLENIC ACID, Fatty acid-binding protein, ... | | Authors: | Sugiyama, S, Nakano, R, Matsuoka, S, Tsuchikawa, H, Sonoyama, M, Inoue, Y, Hayashi, F, Murata, M. | | Deposit date: | 2021-07-24 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (0.88 Å) | | Cite: | The 0.88 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with alpha-llinolenic acid
To Be Published
|
|
7FG1
 
 | | The 0.93 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with tridecanoic acid | | Descriptor: | Fatty acid-binding protein, heart, HEXAETHYLENE GLYCOL, ... | | Authors: | Sugiyama, S, Takahashi, J, Matsuoka, S, Tsuchikawa, H, Sonoyama, M, Inoue, Y, Hayashi, F, Murata, M. | | Deposit date: | 2021-07-25 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (0.93 Å) | | Cite: | The 0.93 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with tridecanoic acid
To Be Published
|
|
3MXO
 
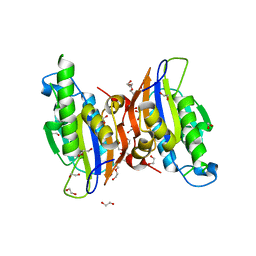 | | Crystal structure oh human phosphoglycerate mutase family member 5 (PGAM5) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Chaikuad, A, Alfano, I, Picaud, S, Filippakopoulos, P, Barr, A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Takeda, K, Ichijo, H, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-05-07 | | Release date: | 2010-09-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of PGAM5 Provide Insight into Active Site Plasticity and Multimeric Assembly.
Structure, 25, 2017
|
|
4MT7
 
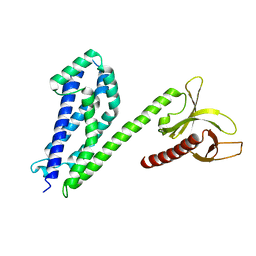 | | Crystal structure of collybistin I | | Descriptor: | Rho guanine nucleotide exchange factor 9 | | Authors: | Schneeberger, D, Schindelin, H. | | Deposit date: | 2013-09-19 | | Release date: | 2014-08-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | A conformational switch in collybistin determines the differentiation of inhibitory postsynapses.
Embo J., 33, 2014
|
|
2D3M
 
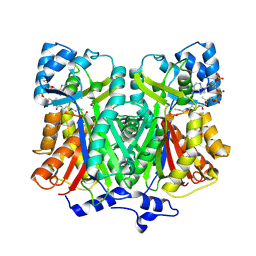 | | Pentaketide chromone synthase complexed with coenzyme A | | Descriptor: | COENZYME A, pentaketide chromone synthase | | Authors: | Morita, H, Kondo, S, Oguro, S, Noguchi, H, Sugio, S, Abe, I, Kohno, T. | | Deposit date: | 2005-09-29 | | Release date: | 2006-10-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Insight into Chain-Length Control and Product Specificity of Pentaketide Chromone Synthase from Aloe arborescens
Chem.Biol., 14, 2007
|
|
5Y3T
 
 | | Crystal structure of hetero-trimeric core of LUBAC: HOIP double-UBA complexed with HOIL-1L UBL and SHARPIN UBL | | Descriptor: | E3 ubiquitin-protein ligase RNF31, GLYCEROL, RanBP-type and C3HC4-type zinc finger-containing protein 1, ... | | Authors: | Tokunaga, A, Fujita, H, Ariyoshi, M, Ohki, I, Tochio, H, Iwai, K, Shirakawa, M. | | Deposit date: | 2017-07-31 | | Release date: | 2018-05-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Cooperative Domain Formation by Homologous Motifs in HOIL-1L and SHARPIN Plays A Crucial Role in LUBAC Stabilization.
Cell Rep, 23, 2018
|
|
4OYU
 
 | | Crystal structure of the N-terminal domains of muskelin | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Muskelin | | Authors: | Delto, C, Kuper, J, Schindelin, H. | | Deposit date: | 2014-02-13 | | Release date: | 2015-02-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The LisH Motif of Muskelin Is Crucial for Oligomerization and Governs Intracellular Localization.
Structure, 23, 2015
|
|
4PD0
 
 | |
