8Z8N
 
 | | Cryo-EM structure of Thogoto virus polymerase in transcription pre-initiation conformation 3 | | Descriptor: | Polymerase acidic protein, Polymerase basic protein 2, RNA (5'-R(*AP*GP*AP*GP*AP*AP*AP*UP*CP*AP*AP*GP*GP*CP*AP*GP*UP*U)-3'), ... | | Authors: | Xue, L, Chang, T, Li, Z, Zhao, H, Li, M, He, J, Chen, X, Xiong, X. | | Deposit date: | 2024-04-22 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Cryo-EM structures of Thogoto virus polymerase reveal unique RNA transcription and replication mechanisms among orthomyxoviruses.
Nat Commun, 15, 2024
|
|
8Z97
 
 | | Cryo-EM structure of Thogoto virus polymerase in a transcription elongation conformation | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-(2'-O-METHYL)-ADENOSINE, Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Xue, L, Chang, T, Li, Z, Zhao, H, Li, M, He, J, Chen, X, Xiong, X. | | Deposit date: | 2024-04-22 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Cryo-EM structures of Thogoto virus polymerase reveal unique RNA transcription and replication mechanisms among orthomyxoviruses.
Nat Commun, 15, 2024
|
|
8Z9H
 
 | | Cryo-EM structure of Thogoto virus polymerase in a transcription elongation-reception conformation | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-(2'-O-METHYL)-ADENOSINE, Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Xue, L, Chang, T, Li, Z, Zhao, H, Li, M, He, J, Chen, X, Xiong, X. | | Deposit date: | 2024-04-23 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM structures of Thogoto virus polymerase reveal unique RNA transcription and replication mechanisms among orthomyxoviruses.
Nat Commun, 15, 2024
|
|
8Z9R
 
 | | Cryo-EM structure of Thogoto virus polymerase in a replication elongation-reception conformation | | Descriptor: | Polymerase acidic protein, Polymerase basic protein 2, RNA (5'-D(*(ATP))-R(P*GP*CP*AP*AP*AP*AP*AP*CP*A)-3'), ... | | Authors: | Xue, L, Chang, T, Li, Z, Zhao, H, Li, M, He, J, Chen, X, Xiong, X. | | Deposit date: | 2024-04-23 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.58 Å) | | Cite: | Cryo-EM structures of Thogoto virus polymerase reveal unique RNA transcription and replication mechanisms among orthomyxoviruses.
Nat Commun, 15, 2024
|
|
8Z8J
 
 | | Cryo-EM structure of Thogoto virus polymerase in transcription pre-initiation conformation 2 | | Descriptor: | Polymerase acidic protein, Polymerase basic protein 2, RNA (5'-R(*AP*GP*AP*GP*AP*AP*AP*UP*CP*AP*AP*GP*GP*CP*AP*GP*UP*U)-3'), ... | | Authors: | Xue, L, Chang, T, Li, Z, Zhao, H, Li, M, He, J, Chen, X, Xiong, X. | | Deposit date: | 2024-04-22 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Cryo-EM structures of Thogoto virus polymerase reveal unique RNA transcription and replication mechanisms among orthomyxoviruses.
Nat Commun, 15, 2024
|
|
8Z9Q
 
 | | Cryo-EM structure of Thogoto virus polymerase in a replication reception conformation | | Descriptor: | Polymerase acidic protein, Polymerase basic protein 2, RNA (5'-D(*(ATP))-R(P*GP*CP*AP*AP*AP*AP*AP*CP*A)-3'), ... | | Authors: | Xue, L, Chang, T, Li, Z, Zhao, H, Li, M, He, J, Chen, X, Xiong, X. | | Deposit date: | 2024-04-23 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.33 Å) | | Cite: | Cryo-EM structures of Thogoto virus polymerase reveal unique RNA transcription and replication mechanisms among orthomyxoviruses.
Nat Commun, 15, 2024
|
|
8Z8X
 
 | | Cryo-EM structure of Thogoto virus polymerase in a transcription initiation conformation | | Descriptor: | PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Xue, L, Chang, T, Li, Z, Zhao, H, Li, M, He, J, Chen, X, Xiong, X. | | Deposit date: | 2024-04-22 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Cryo-EM structures of Thogoto virus polymerase reveal unique RNA transcription and replication mechanisms among orthomyxoviruses.
Nat Commun, 15, 2024
|
|
8Z85
 
 | | Cryo-EM structure of Thogoto virus polymerase in transcription pre-initiation conformation 1 | | Descriptor: | Polymerase acidic protein, Polymerase basic protein 2, RNA (5'-R(*AP*GP*AP*GP*AP*AP*AP*UP*CP*AP*AP*GP*GP*CP*AP*GP*UP*U)-3'), ... | | Authors: | Xue, L, Chang, T, Li, Z, Zhao, H, Li, M, He, J, Chen, X, Xiong, X. | | Deposit date: | 2024-04-21 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Cryo-EM structures of Thogoto virus polymerase reveal unique RNA transcription and replication mechanisms among orthomyxoviruses.
Nat Commun, 15, 2024
|
|
8HHG
 
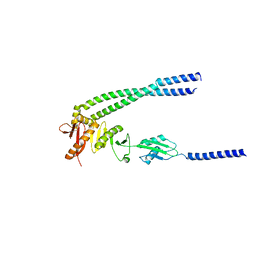 | | The bacterial divisome protein complex FtsB-FtsL-FtsQ | | Descriptor: | Cell division protein FtsB, Cell division protein FtsL, Cell division protein FtsQ | | Authors: | Nguyen, V.H.T, Chen, X. | | Deposit date: | 2022-11-16 | | Release date: | 2023-04-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the heterotrimeric membrane protein complex FtsB-FtsL-FtsQ of the bacterial divisome.
Nat Commun, 14, 2023
|
|
8HHH
 
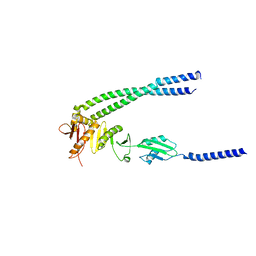 | | The bacterial divisome protein complex FtsB-FtsL-FtsQ | | Descriptor: | Cell division protein FtsB, Cell division protein FtsL, Cell division protein FtsQ | | Authors: | Nguyen, V.H.T, Chen, X. | | Deposit date: | 2022-11-16 | | Release date: | 2023-04-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of the heterotrimeric membrane protein complex FtsB-FtsL-FtsQ of the bacterial divisome.
Nat Commun, 14, 2023
|
|
8HHF
 
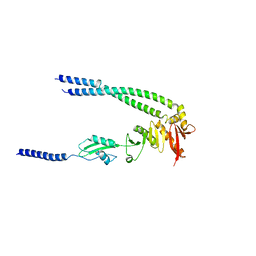 | | The bacterial divisome protein complex FtsB-FtsL-FtsQ | | Descriptor: | Cell division protein FtsB, Cell division protein FtsL, Cell division protein FtsQ | | Authors: | Nguyen, V.H.T, Chen, X. | | Deposit date: | 2022-11-16 | | Release date: | 2023-04-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Structure of the heterotrimeric membrane protein complex FtsB-FtsL-FtsQ of the bacterial divisome.
Nat Commun, 14, 2023
|
|
7CN9
 
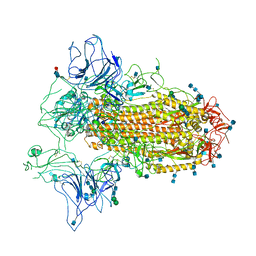 | | Cryo-EM structure of SARS-CoV-2 Spike ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ho, M, Chang, Y, Wang, C, Wu, Y, Huang, H, Chen, T, Lo, J.M, Chen, X, Ma, C. | | Deposit date: | 2020-07-30 | | Release date: | 2020-08-26 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | A Carbohydrate-Binding Protein from the Edible Lablab Beans Effectively Blocks the Infections of Influenza Viruses and SARS-CoV-2.
Cell Rep, 32, 2020
|
|
7YCK
 
 | | Crystal structure of SARS-CoV-2 Spike RBD in complex with FP-12A Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, FP-12A Fab heavy chain, FP-12A Fab light chain, ... | | Authors: | Nguyen, V.H.T, Chen, X. | | Deposit date: | 2022-07-01 | | Release date: | 2023-02-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for a conserved neutralization epitope on the receptor-binding domain of SARS-CoV-2.
Nat Commun, 14, 2023
|
|
7YCL
 
 | | Crystal structure of SARS-CoV-2 Spike RBD in complex with IS-9A Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, IS-9A Fab heavy chain, IS-9A Fab light chain, ... | | Authors: | Mohapatra, A, Chen, X. | | Deposit date: | 2022-07-01 | | Release date: | 2023-02-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structural basis for a conserved neutralization epitope on the receptor-binding domain of SARS-CoV-2.
Nat Commun, 14, 2023
|
|
7YCN
 
 | |
8JQJ
 
 | | Crystal structure of carbonyl reductase SSCR mutant 1 from Sporobolomyces Salmonicolor | | Descriptor: | Aldehyde reductase 2 | | Authors: | Zhang, H.L, Li, Q, Liu, W.D, Chen, X, Wu, Q.Q, Zhu, D.M. | | Deposit date: | 2023-06-14 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Engineering a Carbonyl Reductase to Simultaneously Increase Activity Toward Bulky Ketone and Isopropanol for Dynamic Kinetic Asymmetric Reduction via Enzymatic Hydrogen Transfer
Acs Catalysis, 13, 2023
|
|
8JQK
 
 | | Crystal structure of a carbonyl reductase SSCR mutant from Sporobolomyces Salmonicolor | | Descriptor: | Aldehyde reductase 2 | | Authors: | Zhang, H.L, Li, Q, Liu, W.D, Chen, X, Wu, Q.Q, Zhu, D.M. | | Deposit date: | 2023-06-14 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Engineering a Carbonyl Reductase to Simultaneously Increase Activity Toward Bulky Ketone and Isopropanol for Dynamic Kinetic Asymmetric Reduction via Enzymatic Hydrogen Transfer
Acs Catalysis, 13, 2023
|
|
8YD4
 
 | | CryoEM structure of apo M. tuberculosis ClpP1P2 | | Descriptor: | ATP-dependent Clp protease proteolytic subunit 1, ATP-dependent Clp protease proteolytic subunit 2 | | Authors: | Zhou, B, Gao, Y, Zhao, H, Chen, X, He, J, Zhang, T, Xiong, X. | | Deposit date: | 2024-02-19 | | Release date: | 2025-02-12 | | Method: | ELECTRON MICROSCOPY (3.69 Å) | | Cite: | Activation mechanism of caseinolytic chaperone-protease system in Mycobacterium tuberculosis by the anti-cancer drug bortezomib
To Be Published
|
|
8YCX
 
 | | CryoEM structure of M. tuberculosis ClpC1P1P2 complex bound to bortezomib, conformation 2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpC1, ... | | Authors: | Zhou, B, Zhao, H, Gao, Y, Chen, X, Zhang, T, He, J, Xiong, X. | | Deposit date: | 2024-02-18 | | Release date: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Activation mechanism of caseinolytic chaperone-protease system in Mycobacterium tuberculosis by the anti-cancer drug bortezomib
To Be Published
|
|
8YD0
 
 | | CryoEM structure of M. tuberculosis ClpXP1P2 complex bound to bortezomib | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpX, ... | | Authors: | Zhou, B, Zhao, H, Gao, Y, Chen, X, Zhang, T, He, J, Xiong, X. | | Deposit date: | 2024-02-18 | | Release date: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (2.24 Å) | | Cite: | Activation mechanism of caseinolytic chaperone-protease system in Mycobacterium tuberculosis by the anti-cancer drug bortezomib
To Be Published
|
|
8YD1
 
 | | CryoEM structure of M. tuberculosis ClpC1P1P2 complex bound to bortezomib, conformation 1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpC1, ... | | Authors: | Zhou, B, Zhao, H, Gao, Y, Chen, X, Zhang, T, He, J, Xiong, X. | | Deposit date: | 2024-02-19 | | Release date: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Activation mechanism of caseinolytic chaperone-protease system in Mycobacterium tuberculosis by the anti-cancer drug bortezomib
To Be Published
|
|
7WLT
 
 | | the Curved Structure of mPIEZO1 in Lipid Bilayer | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, O-[(R)-{[(2R)-2,3-bis(octadecanoyloxy)propyl]oxy}(hydroxy)phosphoryl]-L-serine, ... | | Authors: | Yang, X, Lin, C, Chen, X, Li, S, Li, X, Xiao, B. | | Deposit date: | 2022-01-13 | | Release date: | 2022-04-13 | | Last modified: | 2022-07-06 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Structure deformation and curvature sensing of PIEZO1 in lipid membranes.
Nature, 604, 2022
|
|
7WLU
 
 | | The Flattened Structure of mPIEZO1 in Lipid Bilayer | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, Piezo-type mechanosensitive ion channel component 1 | | Authors: | Yang, X, Lin, C, Chen, X, Li, S, Li, X, Xiao, B. | | Deposit date: | 2022-01-13 | | Release date: | 2022-04-13 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (6.81 Å) | | Cite: | Structure deformation and curvature sensing of PIEZO1 in lipid membranes.
Nature, 604, 2022
|
|
5ZHY
 
 | | Structural characterization of the HCoV-229E fusion core | | Descriptor: | Spike glycoprotein | | Authors: | Zhang, W, Zheng, Q, Yan, M, Chen, X, Yang, H, Zhou, W, Rao, Z. | | Deposit date: | 2018-03-13 | | Release date: | 2018-06-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.441 Å) | | Cite: | Structural characterization of the HCoV-229E fusion core.
Biochem. Biophys. Res. Commun., 497, 2018
|
|
3NVN
 
 | | Molecular mechanism of guidance cue recognition | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Liu, H, Juo, Z, Shim, A, Focia, P, Chen, X, Garcia, C, He, X. | | Deposit date: | 2010-07-08 | | Release date: | 2010-09-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structural Basis of Semaphorin-Plexin Recognition and Viral Mimicry from Sema7A and A39R Complexes with PlexinC1.
Cell(Cambridge,Mass.), 142, 2010
|
|
