6QVJ
 
 | | HsCKK (human CAMSAP1) decorated 14pf taxol-GDP microtubule | | Descriptor: | Calmodulin-regulated spectrin-associated protein 1, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Atherton, J.M, Luo, Y, Xiang, S, Yang, C, Jiang, K, Stangier, M, Vemu, A, Cook, A, Wang, S, Roll-Mecak, A, Steinmetz, M.O, Akhmanova, A, Baldus, M, Moores, C.A. | | Deposit date: | 2019-03-02 | | Release date: | 2019-11-27 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural determinants of microtubule minus end preference in CAMSAP CKK domains.
Nat Commun, 10, 2019
|
|
2V4M
 
 | | The isomerase domain of human glutamine-fructose-6-phosphate transaminase 1 (GFPT1) in complex with fructose 6-phosphate | | Descriptor: | CHLORIDE ION, FRUCTOSE -6-PHOSPHATE, GLUCOSAMINE--FRUCTOSE-6-PHOSPHATE AMINOTRANSFERASE [ISOMERIZING] 1 | | Authors: | Moche, M, Lehtio, L, Andersson, J, Arrowsmith, C.H, Berglund, H, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Karlberg, T, Kotenyova, T, Nilsson, M.E, Nyman, T, Persson, C, Sagemark, J, Svensson, S, Schueler, H, Thorsell, A.G, Tresaugues, L, Uppenberg, J, Van Den Berg, S, Welin, M, Wisniewska, M, Weigelt, J, Nordlund, P, Wikstrom, M. | | Deposit date: | 2008-09-26 | | Release date: | 2008-10-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | The Isomerase Domain of Human Gfpt1 in Complex with Fructose 6-Phosphate
To be Published
|
|
7BHX
 
 | | Crystal structure of MAT2a bound to allosteric inhibitor (compound 31) | | Descriptor: | 7-chloranyl-4-(dimethylamino)-1-pyridin-3-yl-quinazolin-2-one, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Schimpl, M, De Fusco, C, Borjesson, U, Cheung, T, Collie, I, Evans, L, Narasimhan, P, Stubbs, C, Vazquez-Chantada, M, Wagner, D.J, Grondine, M, Tentarelli, S, Underwood, E, Argyrou, A, Bagal, S, Chiarparin, E, Robb, G, Scott, J.S. | | Deposit date: | 2021-01-11 | | Release date: | 2021-04-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Fragment-Based Design of a Potent MAT2a Inhibitor and in Vivo Evaluation in an MTAP Null Xenograft Model.
J.Med.Chem., 64, 2021
|
|
3N9V
 
 | | Crystal Structure of INPP5B | | Descriptor: | CALCIUM ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Tresaugues, L, Welin, M, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Karlberg, T, Kol, S, Kotenyova, T, Moche, M, Nyman, T, Persson, C, Schuler, H, Schutz, P, Siponen, M.I, Thorsell, A.G, van der Berg, S, Wahlberg, E, Weigelt, J, Wisniewska, M, Nordlund, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-05-31 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural basis for phosphoinositide substrate recognition, catalysis, and membrane interactions in human inositol polyphosphate 5-phosphatases
Structure, 22, 2014
|
|
3NDH
 
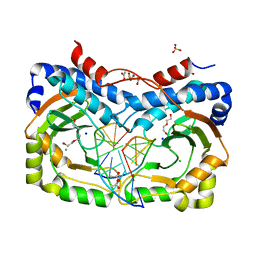 | | Restriction endonuclease in complex with substrate DNA | | Descriptor: | ACETATE ION, CHLORIDE ION, DNA (5'-D(*C*CP*AP*TP*CP*GP*CP*GP*TP*AP*C)-3'), ... | | Authors: | Firczuk, M, Wojciechowski, M, Czapinska, H, Bochtler, M. | | Deposit date: | 2010-06-07 | | Release date: | 2010-07-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | DNA intercalation without flipping in the specific ThaI-DNA complex
Nucleic Acids Res., 39, 2011
|
|
7ZPK
 
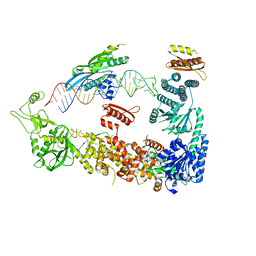 | | Mammalian Dicer in the "pre-dicing state" with pre-miR-15a substrate and TARBP2 subunit | | Descriptor: | 59-nt precursor of miR-15a, Endoribonuclease Dicer, RISC-loading complex subunit TARBP2 | | Authors: | Zanova, M, Zapletal, D, Kubicek, K, Stefl, R, Pinkas, M, Novacek, J. | | Deposit date: | 2022-04-27 | | Release date: | 2022-11-16 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.81 Å) | | Cite: | Structural and functional basis of mammalian microRNA biogenesis by Dicer.
Mol.Cell, 82, 2022
|
|
5LH7
 
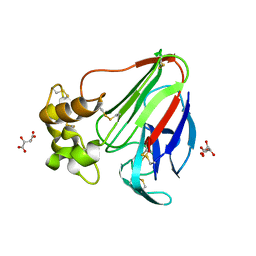 | | High dose Thaumatin - 760-800 ms. | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin-1 | | Authors: | Schubert, R, Kapis, S, Heymann, M, Giquel, Y, Bourenkov, G, Schneider, T, Betzel, C, Perbandt, M. | | Deposit date: | 2016-07-08 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | A multicrystal diffraction data-collection approach for studying structural dynamics with millisecond temporal resolution.
IUCrJ, 3, 2016
|
|
6ZG0
 
 | | SARM1 SAM1-2 domains | | Descriptor: | 1,2-ETHANEDIOL, BETA-MERCAPTOETHANOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sporny, M, Guez-Haddad, J, Khazma, T, Yaron, A, Dessau, M, Mim, C, Isupov, M.N, Zalk, R, Hons, M, Opatowsky, Y. | | Deposit date: | 2020-06-18 | | Release date: | 2020-11-11 | | Last modified: | 2020-12-09 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Structural basis for SARM1 inhibition and activation under energetic stress.
Elife, 9, 2020
|
|
7ZPI
 
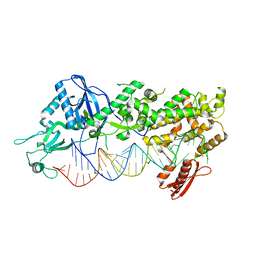 | | Mammalian Dicer in the "dicing state" with pre-miR-15a substrate | | Descriptor: | 59-nt precursor of miR-15a, Endoribonuclease Dicer | | Authors: | Zanova, M, Zapletal, D, Kubicek, K, Stefl, R, Pinkas, M, Novacek, J. | | Deposit date: | 2022-04-27 | | Release date: | 2022-11-16 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (5.91 Å) | | Cite: | Structural and functional basis of mammalian microRNA biogenesis by Dicer.
Mol.Cell, 82, 2022
|
|
5LN0
 
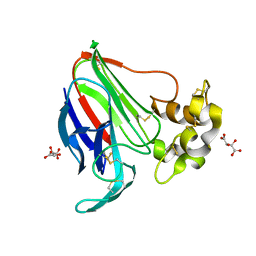 | | Low dose Thaumatin - 760-800 ms. | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin-1 | | Authors: | Schubert, R, Kapis, S, Heymann, M, Giquel, Y, Bourenkov, G, Schneider, T, Betzel, C, Perbandt, M. | | Deposit date: | 2016-08-02 | | Release date: | 2016-11-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A multicrystal diffraction data-collection approach for studying structural dynamics with millisecond temporal resolution.
IUCrJ, 3, 2016
|
|
5LKD
 
 | | Crystal structure of the Xi glutathione transferase ECM4 from Saccharomyces cerevisiae in complex with glutathione | | Descriptor: | GLUTATHIONE, Glutathione S-transferase omega-like 2 | | Authors: | Schwartz, M, Didierjean, C, Hecker, A, Girardet, J.M, Morel-Rouhier, M, Gelhaye, E, Favier, F. | | Deposit date: | 2016-07-22 | | Release date: | 2016-10-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal Structure of Saccharomyces cerevisiae ECM4, a Xi-Class Glutathione Transferase that Reacts with Glutathionyl-(hydro)quinones.
Plos One, 11, 2016
|
|
3NFK
 
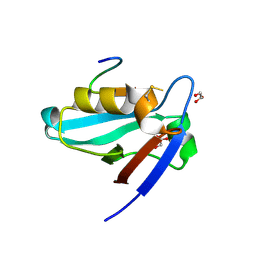 | | Crystal structure of the PTPN4 PDZ domain complexed with the C-terminus of a rabies virus G protein | | Descriptor: | GLYCEROL, Glycoprotein G, Tyrosine-protein phosphatase non-receptor type 4 | | Authors: | Babault, N, Cordier, F, Lafage, M, Cockburn, J, Haouz, A, Rey, F.A, Delepierre, M, Buc, H, Lafon, M, Wolff, N. | | Deposit date: | 2010-06-10 | | Release date: | 2011-08-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Peptides Targeting the PDZ Domain of PTPN4 Are Efficient Inducers of Glioblastoma Cell Death.
Structure, 19, 2011
|
|
5LFJ
 
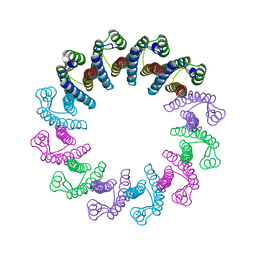 | | Crystal Structure of the Bacterial Proteasome Activator Bpa of Mycobacterium tuberculosis | | Descriptor: | Bacterial proteasome activator | | Authors: | Bolten, M, Delley, C.L, Leibundgut, M, Boehringer, D, Ban, N, Weber-Ban, E. | | Deposit date: | 2016-07-01 | | Release date: | 2016-11-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Analysis of the Bacterial Proteasome Activator Bpa in Complex with the 20S Proteasome.
Structure, 24, 2016
|
|
8BWU
 
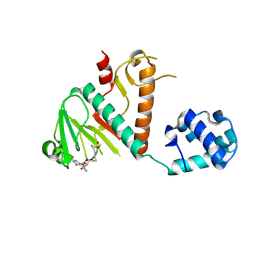 | | Crystal structure of SARS-CoV-2 nsp14 methyltransferase domain in complex with the SS148 inhibitor | | Descriptor: | (2~{S})-2-azanyl-4-[[(2~{S},3~{S},4~{R},5~{R})-5-(4-azanyl-5-cyano-pyrrolo[2,3-d]pyrimidin-7-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanyl]butanoic acid, Transcription factor ETV6,Proofreading exoribonuclease nsp14, ZINC ION | | Authors: | Konkolova, E, Klima, M, Boura, E, Jin, J, Kaniskan, H.U, Han, Y, Vedadi, M. | | Deposit date: | 2022-12-07 | | Release date: | 2023-10-11 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Application of established computational techniques to identify potential SARS-CoV-2 Nsp14-MTase inhibitors in low data regimes
Digit Discov, 2024
|
|
7A22
 
 | | Crystal structure of human protein kinase CK2alpha' (CSNK2A2 gene product) in complex with the ATP-competitive inhibitor 5,6,7-tribromo-1H-triazolo[4,5-b]pyridine | | Descriptor: | 1,2-ETHANEDIOL, 5,6,7-tris(bromanyl)-1~{H}-[1,2,3]triazolo[4,5-b]pyridine, Casein kinase II subunit alpha' | | Authors: | Niefind, K, Lindenblatt, D, Toelzer, C, Bretner, M, Chojnacki, K, Wielechowska, M, Winska, P. | | Deposit date: | 2020-08-15 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.01 Å) | | Cite: | Synthesis, biological properties and structural study of new halogenated azolo[4,5-b]pyridines as inhibitors of CK2 kinase.
Bioorg.Chem., 106, 2021
|
|
7A4B
 
 | | Crystal structure of human protein kinase CK2alpha (CSNK2A1 gene product) in complex with the ATP-competitive inhibitor 5,6-dibromo-1H-triazolo[4,5-b]pyridine | | Descriptor: | 5,6-dibromo-1H-triazolo[4,5-b]pyridine, Casein kinase II subunit alpha, GLYCEROL, ... | | Authors: | Niefind, K, Lindenblatt, D, Toelzer, C, Bretner, M, Chojnacki, K, Wielechowska, M, Winska, P. | | Deposit date: | 2020-08-19 | | Release date: | 2020-12-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Synthesis, biological properties and structural study of new halogenated azolo[4,5-b]pyridines as inhibitors of CK2 kinase.
Bioorg.Chem., 106, 2021
|
|
5JJ8
 
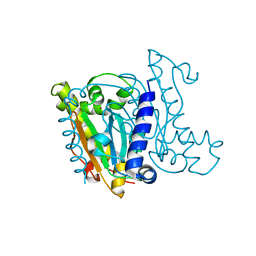 | | Crystal Structure of the Beta Carbonic Anhydrase psCA3 isolated from Pseudomonas aeruginosa - alternate crystal packing form | | Descriptor: | Carbonic anhydrase, ZINC ION | | Authors: | Pinard, M.A, Kurian, J.J, Aggarwal, M, Agbandje-McKenna, M, McKenna, R. | | Deposit date: | 2016-04-22 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.585 Å) | | Cite: | Cryoannealing-induced space-group transition of crystals of the carbonic anhydrase psCA3.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
4U8U
 
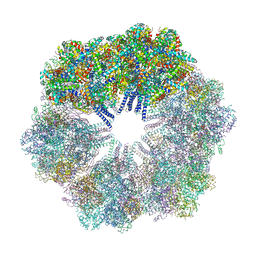 | | The Crystallographic structure of the giant hemoglobin from Glossoscolex paulistus at 3.2 A resolution. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CYANIDE ION, ... | | Authors: | Bachega, J.F.R, Maluf, F.V, Andi, B, D'Muniz Pereira, H, Carazzollea, M.F, Orville, A, Tabak, M, Garratt, R.C, Horjales, E. | | Deposit date: | 2014-08-04 | | Release date: | 2015-06-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The structure of the giant haemoglobin from Glossoscolex paulistus.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3FV1
 
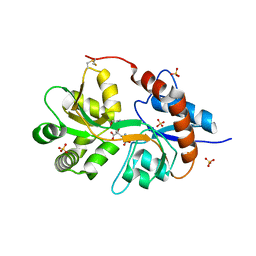 | | Crystal Structure of the human glutamate receptor, GluR5, ligand-binding core in complex with dysiherbaine in space group P1 | | Descriptor: | (2R,3aR,6S,7R,7aR)-2-[(2S)-2-amino-2-carboxyethyl]-6-hydroxy-7-(methylamino)hexahydro-2H-furo[3,2-b]pyran-2-carboxylic acid, GLYCEROL, Glutamate receptor, ... | | Authors: | Unno, M, Sasaki, M, Ikeda-Saito, M. | | Deposit date: | 2009-01-15 | | Release date: | 2010-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Binding and Selectivity of the Marine Toxin Neodysiherbaine A and Its Synthetic Analogues to GluK1 and GluK2 Kainate Receptors.
J.Mol.Biol., 413, 2011
|
|
7VHD
 
 | | Crystal structure of the STX2a complexed with R4A peptide | | Descriptor: | 3-PYRIDINIUM-1-YLPROPANE-1-SULFONATE, ARG-ARG-ARG-ARG-ALA, Shiga toxin 2 B subunit, ... | | Authors: | Senda, M, Takahashi, M, Nishikawa, K, Senda, T. | | Deposit date: | 2021-09-22 | | Release date: | 2022-07-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A unique peptide-based pharmacophore identifies an inhibitory compound against the A-subunit of Shiga toxin.
Sci Rep, 12, 2022
|
|
6OTD
 
 | | Globin sensor domain of AfGcHK in monomeric form, with imidazole | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Globin-coupled histidine kinase, IMIDAZOLE, ... | | Authors: | Skalova, T, Dohnalek, J, Kolenko, P, Stranava, M, Lengalova, A, Martinkova, M. | | Deposit date: | 2019-05-03 | | Release date: | 2020-01-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Disruption of the dimerization interface of the sensing domain in the dimeric heme-based oxygen sensorAfGcHK abolishes bacterial signal transduction.
J.Biol.Chem., 295, 2020
|
|
5JLV
 
 | | Receptor binding domain of Botulinum neurotoxin A in complex with human glycosylated SV2C | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Yao, G, Zhang, S, Mahrhold, S, Lam, K, Stern, D, Bagramyan, K, Perry, K, Kalkum, M, Rummel, A, Dong, M, Jin, R. | | Deposit date: | 2016-04-27 | | Release date: | 2016-06-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | N-linked glycosylation of SV2 is required for binding and uptake of botulinum neurotoxin A.
Nat.Struct.Mol.Biol., 23, 2016
|
|
6ZPR
 
 | | Solution structure of MLKL executioner domain in complex with a covalent inhibitor | | Descriptor: | 7-(2-methoxyethoxymethyl)-1,3-dimethyl-purine-2,6-dione, Mixed lineage kinase domain-like protein,Mixed lineage kinase domain-like protein | | Authors: | Ruebbelke, M, Bauer, M, Hamilton, J, Binder, F, Nar, H, Zeeb, M. | | Deposit date: | 2020-07-09 | | Release date: | 2020-12-16 | | Last modified: | 2021-01-06 | | Method: | SOLUTION NMR | | Cite: | Locking mixed-lineage kinase domain-like protein in its auto-inhibited state prevents necroptosis.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6OR0
 
 | | Crystal structure of Insulin from Non-merohedrally twinned crystals | | Descriptor: | Insulin chain A, Insulin chain B | | Authors: | Sevvana, M, Ruf, M, Uson, I, Sheldrick, G.M, Herbst-Irmer, R. | | Deposit date: | 2019-04-29 | | Release date: | 2019-12-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Non-merohedral twinning: from minerals to proteins.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
7A2D
 
 | | Structure-function analyses of dual-BON domain protein DolP identifies phospholipid binding as a new mechanism for protein localisation to the cell division site | | Descriptor: | Uncharacterized protein YraP | | Authors: | Bryant, J.A, Morris, F.C, Knowles, T.J, Maderbocus, R, Heinz, E, Boelter, G, Alodaini, D, Colyer, A, Wotherspoon, P.J, Staunton, K.A, Jeeves, M, Browning, D.F, Sevastsyanovich, Y.R, Wells, T.J, Rossiter, A.E, Bavro, V.N, Sridhar, P, Ward, D.G, Chong, Z.S, Goodall, E.C.A, Icke, C, Teo, A, Chng, S.S, Roper, D.I, Lithgow, T, Cunningham, A.F, Banzhaf, M, Overduin, M, Henderson, I.R. | | Deposit date: | 2020-08-17 | | Release date: | 2020-12-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of dual BON-domain protein DolP identifies phospholipid binding as a new mechanism for protein localisation.
Elife, 9, 2020
|
|
