4WWQ
 
 | | Apo structure of the Grb7 SH2 domain | | Descriptor: | Growth factor receptor-bound protein 7, MALONIC ACID | | Authors: | Watson, G.M, Ambaye, N.D, Wilce, M.C, Wilce, J.A. | | Deposit date: | 2014-11-12 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cyclic Peptides Incorporating Phosphotyrosine Mimetics as Potent and Specific Inhibitors of the Grb7 Breast Cancer Target.
J.Med.Chem., 58, 2015
|
|
4X6S
 
 | | Grb7 SH2 domain with phosphotyrosine mimetic inhibitor peptide | | Descriptor: | Growth factor receptor-bound protein 7, Phosphotyrosine mimetic inhibitor peptide G7-TEM1 | | Authors: | Watson, G.M, Panjikar, S, Wilce, M.C, Wilce, J.A. | | Deposit date: | 2014-12-09 | | Release date: | 2015-09-23 | | Last modified: | 2015-10-28 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Cyclic Peptides Incorporating Phosphotyrosine Mimetics as Potent and Specific Inhibitors of the Grb7 Breast Cancer Target.
J.Med.Chem., 58, 2015
|
|
4PPK
 
 | | Crystal structure of eCGP123 T69V variant at pH 7.5 | | Descriptor: | Monomeric Azami Green | | Authors: | Don Paul, C, Traore, D.A.K, Devenish, R.J, Close, D, Bell, T, Bradbury, A, Wilce, M.C.J, Prescott, M. | | Deposit date: | 2014-02-27 | | Release date: | 2015-04-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-Ray Crystal Structure and Properties of Phanta, a Weakly Fluorescent Photochromic GFP-Like Protein.
Plos One, 10, 2015
|
|
4PPJ
 
 | | Crystal structure of Phanta, a weakly fluorescent photochromic GFP-like protein. ON state | | Descriptor: | Monomeric Azami Green | | Authors: | Don Paul, C, Traore, D.A.K, Devenish, R.J, Close, D, Bell, T, Bradbury, A, Wilce, M.C.J, Prescott, M. | | Deposit date: | 2014-02-27 | | Release date: | 2015-04-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-Ray Crystal Structure and Properties of Phanta, a Weakly Fluorescent Photochromic GFP-Like Protein.
Plos One, 10, 2015
|
|
4PPL
 
 | | Crystal structure of eCGP123 H193Q variant at pH 7.5 | | Descriptor: | Monomeric Azami Green | | Authors: | Don Paul, C, Traore, D.A.K, Devenish, R.J, Close, D, Bell, T, Bradbury, A, Wilce, M.C.J, Prescott, M. | | Deposit date: | 2014-02-27 | | Release date: | 2015-04-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-Ray Crystal Structure and Properties of Phanta, a Weakly Fluorescent Photochromic GFP-Like Protein.
Plos One, 10, 2015
|
|
1MMI
 
 | | E. COLI DNA POLYMERASE BETA SUBUNIT | | Descriptor: | DNA polymerase III, beta chain | | Authors: | Oakley, A.J, Prosselkov, P, Wijffels, G, Beck, J.L, Wilce, M.C.J, Dixon, N.E. | | Deposit date: | 2002-09-04 | | Release date: | 2003-09-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.848 Å) | | Cite: | Flexibility revealed by the 1.85 A crystal structure of the beta sliding-clamp subunit of Escherichia coli DNA polymerase III.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1B13
 
 | | CLOSTRIDIUM PASTEURIANUM RUBREDOXIN G10A MUTANT | | Descriptor: | FE (III) ION, PROTEIN (RUBREDOXIN) | | Authors: | Maher, M.J, Guss, J.M, Wilce, M.C.J, Wedd, A.G. | | Deposit date: | 1998-11-26 | | Release date: | 1999-05-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Rubredoxin from Clostridium pasteurianum. Structures of G10A, G43A and G10VG43A mutant proteins. Mutation of conserved glycine 10 to valine causes the 9-10 peptide link to invert.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1B2O
 
 | | CLOSTRIDIUM PASTEURIANUM RUBREDOXIN G10VG43A MUTANT | | Descriptor: | FE (III) ION, PROTEIN (RUBREDOXIN) | | Authors: | Maher, M.J, Guss, J.M, Wilce, M.C.J, Wedd, A.G. | | Deposit date: | 1998-11-30 | | Release date: | 1999-05-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rubredoxin from Clostridium pasteurianum. Structures of G10A, G43A and G10VG43A mutant proteins. Mutation of conserved glycine 10 to valine causes the 9-10 peptide link to invert.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1B2J
 
 | | CLOSTRIDIUM PASTEURIANUM RUBREDOXIN G43A MUTANT | | Descriptor: | FE (III) ION, PROTEIN (RUBREDOXIN) | | Authors: | Maher, M.J, Guss, J.M, Wilce, M.C.J, Wedd, A.G. | | Deposit date: | 1998-11-27 | | Release date: | 1999-05-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Rubredoxin from Clostridium pasteurianum. Structures of G10A, G43A and G10VG43A mutant proteins. Mutation of conserved glycine 10 to valine causes the 9-10 peptide link to invert.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1R0F
 
 | | Gallium-substituted rubredoxin | | Descriptor: | GALLIUM (III) ION, Rubredoxin | | Authors: | Maher, M, Cross, M, Wilce, M.C.J, Guss, J.M, Wedd, A.G. | | Deposit date: | 2003-09-22 | | Release date: | 2004-02-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Metal-substituted derivatives of the rubredoxin from Clostridium pasteurianum.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1R0H
 
 | | cobalt-substituted rubredoxin | | Descriptor: | COBALT (II) ION, Rubredoxin | | Authors: | Maher, M, Cross, M, Wilce, M.C.J, Guss, J.M, Wedd, A.G. | | Deposit date: | 2003-09-22 | | Release date: | 2004-02-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Metal-substituted derivatives of the rubredoxin from Clostridium pasteurianum.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
2QMS
 
 | | Crystal structure of a signaling molecule | | Descriptor: | Growth factor receptor-bound protein 7, SULFATE ION | | Authors: | Porter, C.J, Wilce, M.C, Wilce, J.A. | | Deposit date: | 2007-07-16 | | Release date: | 2008-07-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Grb7 SH2 domain structure and interactions with a cyclic peptide inhibitor of cancer cell migration and proliferation.
Bmc Struct.Biol., 7, 2007
|
|
1QHQ
 
 | | AURACYANIN, A BLUE COPPER PROTEIN FROM THE GREEN THERMOPHILIC PHOTOSYNTHETIC BACTERIUM CHLOROFLEXUS AURANTIACUS | | Descriptor: | CHLORIDE ION, COPPER (II) ION, PROTEIN (AURACYANIN), ... | | Authors: | Bond, C.S, Blankenship, R.E, Freeman, H.C, Guss, J.M, Maher, M, Selvaraj, F, Wilce, M.C.J, Willingham, K. | | Deposit date: | 1999-05-25 | | Release date: | 2001-03-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of auracyanin, a "blue" copper protein from the green thermophilic photosynthetic bacterium Chloroflexus aurantiacus.
J.Mol.Biol., 306, 2001
|
|
1RH9
 
 | | Family GH5 endo-beta-mannanase from Lycopersicon esculentum (tomato) | | Descriptor: | endo-beta-mannanase | | Authors: | Oakley, A.J, Bourgault, R, Bewley, J.D, Wilce, M.C.J. | | Deposit date: | 2003-11-14 | | Release date: | 2005-04-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Three-dimensional structure of (1,4)-beta-D-mannan mannanohydrolase from tomato fruit.
Protein Sci., 14, 2005
|
|
1R0G
 
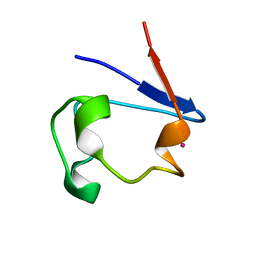 | | mercury-substituted rubredoxin | | Descriptor: | MERCURY (II) ION, Rubredoxin | | Authors: | Maher, M, Cross, M, Wilce, M.C.J, Guss, J.M, Wedd, A.G. | | Deposit date: | 2003-09-22 | | Release date: | 2004-02-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Metal-substituted derivatives of the rubredoxin from Clostridium pasteurianum.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1R0I
 
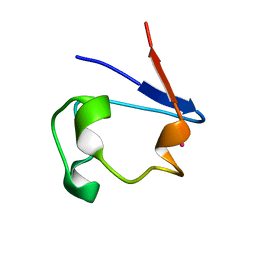 | | cadmium-substituted rubredoxin | | Descriptor: | CADMIUM ION, Rubredoxin | | Authors: | Maher, M, Cross, M, Wilce, M.C.J, Guss, J.M, Wedd, A.G. | | Deposit date: | 2003-09-22 | | Release date: | 2004-02-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Metal-substituted derivatives of the rubredoxin from Clostridium pasteurianum.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1R0J
 
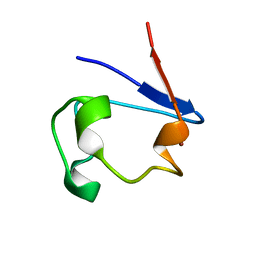 | | nickel-substituted rubredoxin | | Descriptor: | NICKEL (II) ION, Rubredoxin | | Authors: | Maher, M, Cross, M, Wilce, M.C.J, Guss, J.M, Wedd, A.G. | | Deposit date: | 2003-09-22 | | Release date: | 2004-02-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Metal-substituted derivatives of the rubredoxin from Clostridium pasteurianum.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1G4H
 
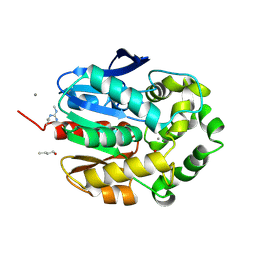 | | LINB COMPLEXED WITH BUTAN-1-OL | | Descriptor: | 1,3,4,6-TETRACHLORO-1,4-CYCLOHEXADIENE HYDROLASE, 1-BUTANOL, CALCIUM ION, ... | | Authors: | Oakley, A.J, Prokop, Z, Bohac, M, Kmunicek, J, Jedlicka, T, Monincova, M, Kuta-Smatanova, I, Nagata, Y, Damborsky, J, Wilce, M.C.J. | | Deposit date: | 2000-10-27 | | Release date: | 2001-10-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Exploring the structure and activity of haloalkane dehalogenase from Sphingomonas paucimobilis UT26: evidence for product- and water-mediated inhibition.
Biochemistry, 41, 2002
|
|
1IGO
 
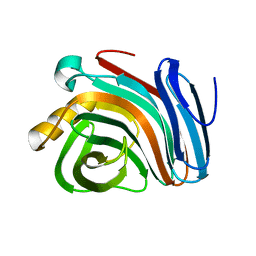 | | Family 11 xylanase | | Descriptor: | SULFATE ION, family 11 xylanase | | Authors: | Oakley, A.J, Thomson, C, Heinrich, T, Dunlop, R, Wilce, M.C.J. | | Deposit date: | 2001-04-18 | | Release date: | 2002-04-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Characterization of a family 11 xylanase from Bacillus subtillis B230 used for paper bleaching.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1G42
 
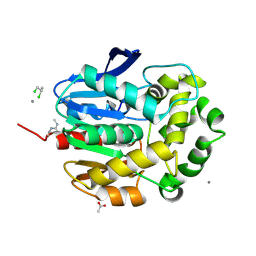 | | STRUCTURE OF 1,3,4,6-TETRACHLORO-1,4-CYCLOHEXADIENE HYDROLASE (LINB) FROM SPHINGOMONAS PAUCIMOBILIS COMPLEXED WITH 1,2-DICHLOROPROPANE | | Descriptor: | 1,2-DICHLORO-PROPANE, 1,3,4,6-TETRACHLORO-1,4-CYCLOHEXADIENE HYDROLASE, ACETATE ION, ... | | Authors: | Oakley, A.J, Prokop, Z, Bohac, M, Kmunicek, J, Jedlicka, T, Monincova, M, Kuta-Smatanova, I, Nagata, Y, Damborsky, J, Wilce, M.C.J. | | Deposit date: | 2000-10-26 | | Release date: | 2001-10-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Exploring the structure and activity of haloalkane dehalogenase from Sphingomonas paucimobilis UT26: evidence for product- and water-mediated inhibition.
Biochemistry, 41, 2002
|
|
1G5F
 
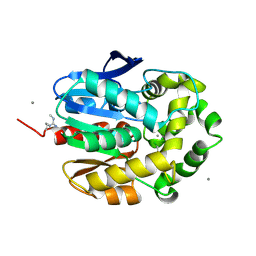 | | STRUCTURE OF LINB COMPLEXED WITH 1,2-DICHLOROETHANE | | Descriptor: | 1,2-DICHLOROETHANE, 1,3,4,6-TETRACHLORO-1,4-CYCLOHEXADIENE HYDROLASE, CALCIUM ION, ... | | Authors: | Oakley, A.J, Prokop, Z, Bohac, M, Kmunicek, J, Jedlicka, T, Monincova, M, Kuta-Smatanova, I, Nagata, Y, Damborsky, J, Wilce, M.C.J. | | Deposit date: | 2000-11-01 | | Release date: | 2001-11-01 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Exploring the structure and activity of haloalkane dehalogenase from Sphingomonas paucimobilis UT26: evidence for product- and water-mediated inhibition.
Biochemistry, 41, 2002
|
|
2AK4
 
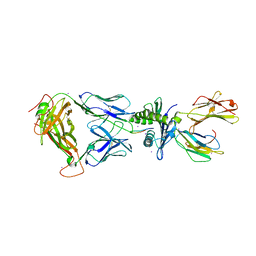 | | Crystal Structure of SB27 TCR in complex with HLA-B*3508-13mer peptide | | Descriptor: | Beta-2-microglobulin, EBV peptide LPEPLPQGQLTAY, HLA-B35 variant, ... | | Authors: | Tynan, F.E, Burrows, S.R, Buckle, A.M, Clements, C.S, Borg, N.A, Miles, J.J, Beddoe, T, Whisstock, J.C, Wilce, M.C, Silins, S.L, Burrows, J.M, Kjer-Nielsen, L, Konstenko, L, Purcell, A.W, McCluskey, J, Rossjohn, J. | | Deposit date: | 2005-08-03 | | Release date: | 2005-10-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | T cell receptor recognition of a 'super-bulged' major histocompatibility complex class I-bound peptide
Nat.Immunol., 6, 2005
|
|
3KPQ
 
 | | Crystal Structure of HLA B*4405 in complex with EEYLKAWTF, a mimotope | | Descriptor: | Beta-2-microglobulin, EEYLKAWTF, mimotope peptide, ... | | Authors: | Macdonald, W.A, Chen, Z, Gras, S, Archbold, J.K, Tynan, F.E, Clements, C.S, Bharadwaj, M, Kjer-Nielsen, L, Saunders, P.M, Wilce, M.C, Crawford, F, Stadinsky, B, Jackson, D, Brooks, A.G, Purcell, A.W, Kappler, J.W, Burrows, S.R, Rossjohn, J, McCluskey, J. | | Deposit date: | 2009-11-16 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | T cell allorecognition via molecular mimicry.
Immunity, 31, 2009
|
|
3KPL
 
 | | Crystal Structure of HLA B*4402 in complex with EEYLQAFTY a self peptide from the ABCD3 protein | | Descriptor: | Beta-2-microglobulin, EEYLQAFTY, self peptide from the ATP binding cassette protein ABCD3, ... | | Authors: | Macdonald, W.A, Chen, Z, Gras, S, Archbold, J.K, Tynan, F.E, Clements, C.S, Bharadwaj, M, Kjer-Nielsen, L, Saunders, P.M, Wilce, M.C, Crawford, F, Stadinsky, B, Jackson, D, Brooks, A.G, Purcell, A.W, Kappler, J.W, Burrows, S.R, Rossjohn, J, McCluskey, J. | | Deposit date: | 2009-11-16 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | T cell allorecognition via molecular mimicry.
Immunity, 31, 2009
|
|
3KPS
 
 | | Crystal Structure of the LC13 TCR in complex with HLA B*4405 bound to EEYLQAFTY a self peptide from the ABCD3 protein | | Descriptor: | Beta-2-microglobulin, EEYLQAFTY, self peptide from the ATP binding cassette protein ABCD3, ... | | Authors: | Macdonald, W.A, Chen, Z, Gras, S, Archbold, J.K, Tynan, F.E, Clements, C.S, Bharadwaj, M, Kjer-Nielsen, L, Saunders, P.M, Wilce, M.C, Crawford, F, Stadinsky, B, Jackson, D, Brooks, A.G, Purcell, A.W, Kappler, J.W, Burrows, S.R, Rossjohn, J, McCluskey, J. | | Deposit date: | 2009-11-16 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | T cell allorecognition via molecular mimicry.
Immunity, 31, 2009
|
|
