3ZKI
 
 | | BACE2 MUTANT STRUCTURE WITH LIGAND | | Descriptor: | 5-(2,2,2-Trifluoro-ethoxy)-pyridine-2-carboxylic acid [3-((S)-2-amino-1,4-dimethyl-6-oxo-1,4,5,6-tetrahydro-pyrimidin-4-yl)-phenyl]-amide, BETA-SECRETASE 2 | | Authors: | Banner, D.W, Kuglstatter, A, Benz, J, Stihle, M, Ruf, A. | | Deposit date: | 2013-01-23 | | Release date: | 2013-05-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mapping the Conformational Space Accessible to Bace2 Using Surface Mutants and Co-Crystals with Fab-Fragments, Fynomers, and Xaperones
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4BEL
 
 | | BACE2 XAPERONE COMPLEX | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-SECRETASE 2, CHLORIDE ION, ... | | Authors: | Banner, D.W, Kuglstatter, A, Benz, J, Stihle, M, Ruf, A. | | Deposit date: | 2013-03-11 | | Release date: | 2013-05-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mapping the Conformational Space Accessible to Bace2 Using Surface Mutants and Co-Crystals with Fab-Fragments, Fynomers, and Xaperones
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4BFB
 
 | | BACE2 XAPERONE COMPLEX | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-SECRETASE 2, XA4813 | | Authors: | Banner, D.W, Kuglstatter, A, Benz, J, Stihle, M, Ruf, A. | | Deposit date: | 2013-03-18 | | Release date: | 2013-05-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Mapping the Conformational Space Accessible to Bace2 Using Surface Mutants and Co-Crystals with Fab-Fragments, Fynomers, and Xaperones
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3ZKQ
 
 | | BACE2 XAPERONE COMPLEX | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-SECRETASE 2, CHLORIDE ION, ... | | Authors: | Banner, D.W, Kuglstatter, A, Benz, J, Stihle, M, Ruf, A. | | Deposit date: | 2013-01-24 | | Release date: | 2013-05-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Mapping the Conformational Space Accessible to Bace2 Using Surface Mutants and Co-Crystals with Fab-Fragments, Fynomers, and Xaperones
Acta Crystallogr.,Sect.D, 69, 2013
|
|
7B0W
 
 | | Crystal structure of the E. coli type 1 pilus assembly inhibitor FimI bound to FimC | | Descriptor: | 1,2-ETHANEDIOL, Chaperone protein FimC, FORMIC ACID, ... | | Authors: | Scharer, M.A, Zigova, Z, Giese, C, Puorger, C, Ignatov, O, Capitani, G, Glockshuber, R. | | Deposit date: | 2020-11-23 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Comprehensive kinetic characterization of bacterial pilus rod assembly and assembly termination
To Be Published
|
|
7B0X
 
 | | Crystal structure of the ternary complex of the E. coli type 1 pilus proteins FimC, FimI and the N-terminal domain of FimD | | Descriptor: | 1,2-ETHANEDIOL, Chaperone protein FimC, Fimbrin-like protein FimI, ... | | Authors: | Scharer, M.A, Zigova, Z, Giese, C, Puorger, C, Ignatov, O, Capitani, G, Glockshuber, R. | | Deposit date: | 2020-11-23 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Comprehensive kinetic characterization of bacterial pilus rod assembly and assembly termination
To Be Published
|
|
6YMC
 
 | | 26-mer stem-loop RNA | | Descriptor: | BARIUM ION, RNA (26-MER) | | Authors: | Janowski, R, Niessing, D. | | Deposit date: | 2020-04-08 | | Release date: | 2022-04-20 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Multiple intrinsically disordered RNA-binding motifs cooperate as RNA-folding catalyst and mediate phase transition
To Be Published
|
|
7Z6L
 
 | | Crystal structure of PROTAC 5 in complex with the bromodomain of human SMARCA2 and pVHL:ElonginC:ElonginB | | Descriptor: | (2~{S},4~{R})-~{N}-[[2-[3-[4-(4-bromanyl-7-cyclopentyl-5-oxidanylidene-benzimidazolo[1,2-a]quinazolin-9-yl)piperidin-1-yl]propoxy]-4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl]-1-[(2~{S})-2-[(1-fluoranylcyclopropyl)carbonylamino]-3,3-dimethyl-butanoyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Elongin-B, Elongin-C, ... | | Authors: | Roy, M.J, Bader, G, Farnaby, W, Ciulli, A. | | Deposit date: | 2022-03-12 | | Release date: | 2022-09-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | A selective and orally bioavailable VHL-recruiting PROTAC achieves SMARCA2 degradation in vivo.
Nat Commun, 13, 2022
|
|
7Z77
 
 | | Crystal structure of compound 6 in complex with the bromodomain of human SMARCA2 and pVHL:ElonginC:ElonginB | | Descriptor: | (2~{S},4~{R})-~{N}-[(1~{S})-4-[4-(4-bromanyl-7-cyclopentyl-5-oxidanylidene-benzimidazolo[1,2-a]quinazolin-9-yl)piperidin-1-yl]-1-[4-(4-methyl-1,3-thiazol-5-yl)phenyl]butyl]-1-[(2~{S})-2-[(1-fluoranylcyclopropyl)carbonylamino]-3,3-dimethyl-butanoyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Elongin-B, Elongin-C, ... | | Authors: | Bader, G, Boettcher, J, Wolkerstorfer, B. | | Deposit date: | 2022-03-15 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | A selective and orally bioavailable VHL-recruiting PROTAC achieves SMARCA2 degradation in vivo.
Nat Commun, 13, 2022
|
|
7Z76
 
 | | Crystal structure of compound 10 in complex with the bromodomain of human SMARCA2 and pVHL:ElonginC:ElonginB | | Descriptor: | (2~{S},4~{R})-~{N}-[(1~{R})-2-[(2~{R})-1-[4-(4-bromanyl-7-cyclopentyl-5-oxidanylidene-benzimidazolo[1,2-a]quinazolin-9-yl)piperidin-1-yl]propan-2-yl]oxy-1-[4-(4-methyl-1,3-thiazol-5-yl)phenyl]ethyl]-1-[(2~{S})-2-[[1-(dimethylamino)cyclopropyl]carbonylamino]-3,3-dimethyl-butanoyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Elongin-B, Elongin-C, ... | | Authors: | Bader, G, Boettcher, J, Wolkerstorfer, B. | | Deposit date: | 2022-03-15 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | A selective and orally bioavailable VHL-recruiting PROTAC achieves SMARCA2 degradation in vivo.
Nat Commun, 13, 2022
|
|
7Z78
 
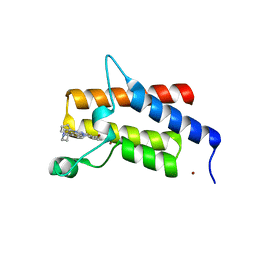 | | Crystal structure of compound 4 in complex with the bromodomain of human SMARCA2 and pVHL:ElonginC:ElonginB | | Descriptor: | 4-bromanyl-7-cyclopentyl-9-piperidin-4-yl-benzimidazolo[1,2-a]quinazolin-5-one, Probable global transcription activator SNF2L2, ZINC ION | | Authors: | Bader, G, Boettcher, J, Wolkerstorfer, B. | | Deposit date: | 2022-03-15 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | A selective and orally bioavailable VHL-recruiting PROTAC achieves SMARCA2 degradation in vivo.
Nat Commun, 13, 2022
|
|
7AFW
 
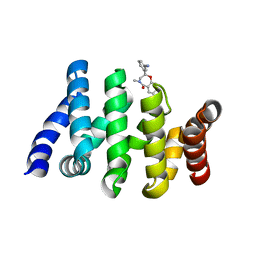 | | Beta-Catenin in complex with compound 6 | | Descriptor: | 3-[(2~{R})-4-methyl-5-oxidanylidene-2,3-dihydro-1,4-benzoxazepin-2-yl]benzenecarbonitrile, Catenin beta-1 | | Authors: | Boettcher, J, Kessler, D. | | Deposit date: | 2020-09-21 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.814 Å) | | Cite: | Getting a Grip on the Undrugged: Targeting beta-Catenin with Fragment-Based Methods.
Chemmedchem, 16, 2021
|
|
5E8C
 
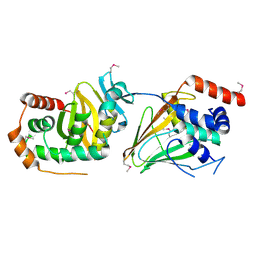 | | pseudorabies virus nuclear egress complex, pUL31, pUL34 | | Descriptor: | CHLORIDE ION, UL31, UL34 protein, ... | | Authors: | Zeev-Ben-Mordehai, T, Cheleski, J, Whittle, C, El Omari, K, Harlos, K, Hagen, C, Klupp, B, Mettenleiter, T.C, Gruenewald, K. | | Deposit date: | 2015-10-14 | | Release date: | 2015-12-23 | | Last modified: | 2016-01-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of the Herpesvirus Nuclear Egress Complex Provides Insights into Inner Nuclear Membrane Remodeling.
Cell Rep, 13, 2015
|
|
5FKI
 
 | | Pseudorabies virus (PrV) nuclear egress complex proteins fitted as a hexameric lattice into a sub-tomogram average derived from focused- ion beam milled lamellae electron cryo-microscopic data | | Descriptor: | CHLORIDE ION, UL31, UL34 protein, ... | | Authors: | Hagen, C, Dent, K.C, Zeev Ben Mordehai, T, Vasishtan, D, Antonin, W, Mettenleiter, T.C, Gruenewald, K. | | Deposit date: | 2015-10-16 | | Release date: | 2016-03-16 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (35 Å) | | Cite: | Crystal Structure of the Herpesvirus Nuclear Egress Complex Provides Insights Into Inner Nuclear Membrane Remodelling
Cell Rep., 13, 2015
|
|
