5KSI
 
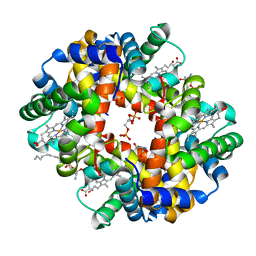 | | Crystal structure of deoxygenated hemoglobin in complex with sphingosine phosphate and 2,3-Bisphosphoglycerate | | Descriptor: | (2R)-2,3-diphosphoglyceric acid, (2S,3R,4E)-2-amino-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, Hemoglobin subunit alpha, ... | | Authors: | Ahmed, M.H, Safo, M.K, Xia, Y. | | Deposit date: | 2016-07-08 | | Release date: | 2017-07-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Functional Insight of Sphingosine 1-Phosphate-Mediated Pathogenic Metabolic Reprogramming in Sickle Cell Disease.
Sci Rep, 7, 2017
|
|
7YH8
 
 | | Crystal structure of a heterochiral protein complex | | Descriptor: | D-Pep-1, L-19437 | | Authors: | Liang, M, Li, S, Wang, T, Liu, L, Lu, P. | | Deposit date: | 2022-07-13 | | Release date: | 2023-07-19 | | Last modified: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Accurate de novo design of heterochiral protein-protein interactions
Cell Res., 2024
|
|
8GQP
 
 | | Complex of D-protein binder D-19437 and L-target L-Pep-1 | | Descriptor: | D-binder, L-pep1 | | Authors: | Liang, M.F, Li, S.C, Wang, T.Y, Liu, L, Lu, P.L. | | Deposit date: | 2022-08-30 | | Release date: | 2023-09-13 | | Last modified: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Accurate de novo design of heterochiral protein-protein interactions
Cell Res., 2024
|
|
6WZZ
 
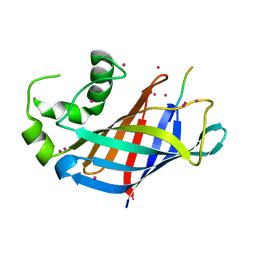 | | GID4 in complex with VGLWKS peptide | | Descriptor: | Glucose-induced degradation protein 4 homolog, UNKNOWN ATOM OR ION, VGLWKS peptide | | Authors: | Dong, C, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-05-14 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Recognition of nonproline N-terminal residues by the Pro/N-degron pathway.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6WZX
 
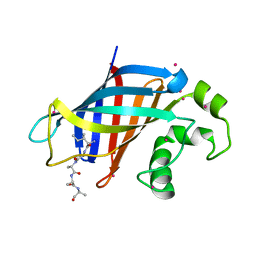 | | GID4 in complex with IGLWKS peptide | | Descriptor: | Glucose-induced degradation protein 4 homolog, ILE-GLY-LEU-TRP-LYS peptide, UNKNOWN ATOM OR ION | | Authors: | Dong, C, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-05-14 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Recognition of nonproline N-terminal residues by the Pro/N-degron pathway.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6L40
 
 | | Discovery of novel peptidomimetic boronate ClpP inhibitors with noncanonical enzyme mechanism as potent virulence blockers in vitro and in vivo | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, [(1S)-3-methyl-1-[[(2S)-3-phenyl-2-(pyrazin-2-ylcarbonylamino)propanoyl]amino]butyl]boronic acid | | Authors: | Luo, Y.F, Bao, R, Ju, Y, He, L.H. | | Deposit date: | 2019-10-15 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.209 Å) | | Cite: | Discovery of Novel Peptidomimetic Boronate ClpP Inhibitors with Noncanonical Enzyme Mechanism as Potent Virulence Blockersin Vitroandin Vivo.
J.Med.Chem., 63, 2020
|
|
6KU0
 
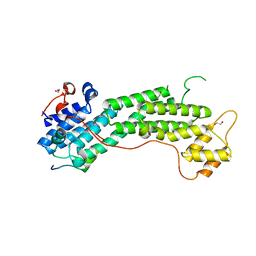 | | Crystal structure of MyoVa-GTD in complex with MICAL1-GTBM | | Descriptor: | 1,2-ETHANEDIOL, Peptide from [F-actin]-monooxygenase MICAL1, Unconventional myosin-Va | | Authors: | Niu, F, Wei, Z. | | Deposit date: | 2019-08-29 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | F-actin disassembly factor MICAL1 binding to Myosin Va mediates cargo unloading during cytokinesis.
Sci Adv, 6, 2020
|
|
6LEA
 
 | |
6L3X
 
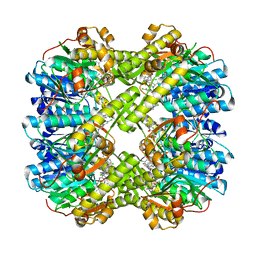 | | Discovery of novel peptidomimetic boronate ClpP inhibitors with noncanonical enzyme mechanism as potent virulence blockers in vitro and in vivo | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, [(1~{R})-1-[[(2~{S})-2-[[2,5-bis(chloranyl)phenyl]carbonylamino]-3-(1~{H}-indol-3-yl)propanoyl]amino]-3-methyl-butyl]boronic acid | | Authors: | Luo, Y.F, Bao, R, Ju, Y, He, L.H. | | Deposit date: | 2019-10-15 | | Release date: | 2020-08-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3054 Å) | | Cite: | Discovery of Novel Peptidomimetic Boronate ClpP Inhibitors with Noncanonical Enzyme Mechanism as Potent Virulence Blockersin Vitroandin Vivo.
J.Med.Chem., 63, 2020
|
|
3BGL
 
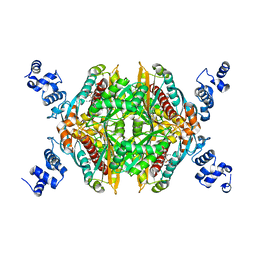 | | Hepatoselectivity of Statins: Design and synthesis of 4-sulfamoyl pyrroles as HMG-CoA reductase inhibitors | | Descriptor: | (3R,5R)-7-[2-(4-fluorophenyl)-5-(1-methylethyl)-4-(morpholin-4-ylsulfonyl)-3-phenyl-1H-pyrrol-1-yl]-3,5-dihydroxyheptanoic acid, 3-hydroxy-3-methylglutaryl-coenzyme A reductase | | Authors: | Finzel, B.C, Pavlovsky, A, Park, W.K.C. | | Deposit date: | 2007-11-26 | | Release date: | 2008-01-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.225 Å) | | Cite: | Hepatoselectivity of statins: design and synthesis of 4-sulfamoyl pyrroles as HMG-CoA reductase inhibitors.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
7YV9
 
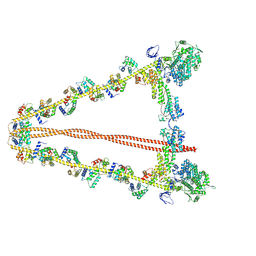 | |
5YAZ
 
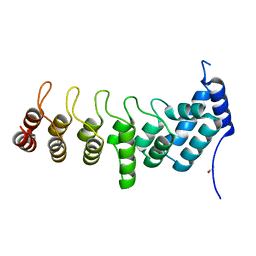 | | Crystal structure of the ANKRD domain of KANK1 | | Descriptor: | ACETATE ION, KN motif and ankyrin repeat domains 1 | | Authors: | Wei, Z, Pan, W. | | Deposit date: | 2017-09-02 | | Release date: | 2017-12-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into ankyrin repeat-mediated recognition of the kinesin motor protein KIF21A by KANK1, a scaffold protein in focal adhesion.
J. Biol. Chem., 293, 2018
|
|
5YAY
 
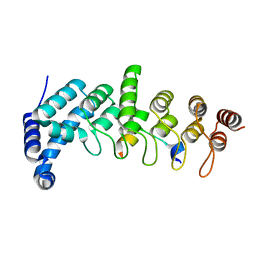 | | Crystal structure of KANK1/KIF21A complex | | Descriptor: | KN motif and ankyrin repeat domains 1, Kinesin-like protein KIF21A | | Authors: | Wei, Z, Pan, W. | | Deposit date: | 2017-09-02 | | Release date: | 2017-12-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural insights into ankyrin repeat-mediated recognition of the kinesin motor protein KIF21A by KANK1, a scaffold protein in focal adhesion.
J. Biol. Chem., 293, 2018
|
|
6BA6
 
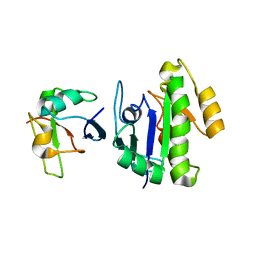 | | Solution structure of Rap1b/talin complex | | Descriptor: | Ras-related protein Rap-1b, Talin-1 | | Authors: | Zhu, L, Yang, J, Qin, J. | | Deposit date: | 2017-10-12 | | Release date: | 2017-12-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of Rap1b bound to talin reveals a pathway for triggering integrin activation.
Nat Commun, 8, 2017
|
|
6LTO
 
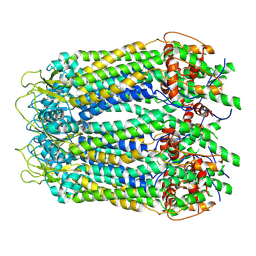 | | cryo-EM structure of full length human Pannexin1 | | Descriptor: | Pannexin-1 | | Authors: | Mou, L.Q, Ke, M, Xiao, Q.J, Wu, J.P, Deng, D. | | Deposit date: | 2020-01-23 | | Release date: | 2020-05-13 | | Last modified: | 2020-05-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for gating mechanism of Pannexin 1 channel.
Cell Res., 30, 2020
|
|
6LTN
 
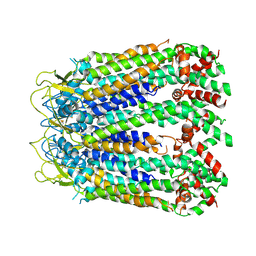 | | cryo-EM structure of C-terminal truncated human Pannexin1 | | Descriptor: | Pannexin-1 | | Authors: | Mou, L.Q, Ke, M, Xiao, Q.J, Wu, J.P, Deng, D. | | Deposit date: | 2020-01-23 | | Release date: | 2020-05-13 | | Last modified: | 2020-05-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for gating mechanism of Pannexin 1 channel.
Cell Res., 30, 2020
|
|
5XRW
 
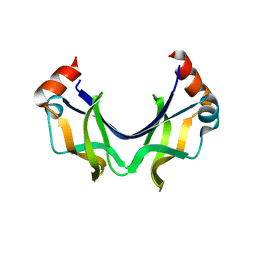 | |
8Y9C
 
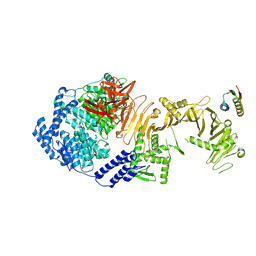 | | De novo design mini-binder in complex with TcdB4 | | Descriptor: | De novo design Minibinder, Toxin B, ZINC ION | | Authors: | Lv, X.C, Lu, P.L. | | Deposit date: | 2024-02-06 | | Release date: | 2024-08-28 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | De novo design of mini-protein binders broadly neutralizing Clostridioides difficile toxin B variants
Nat Commun, 2024
|
|
8Y9B
 
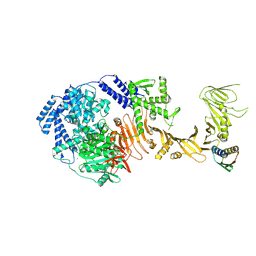 | | TcdB1 in complex with mini-binder | | Descriptor: | De novo design mini-binder, Toxin B, ZINC ION | | Authors: | Lv, X.C, Lu, P.L. | | Deposit date: | 2024-02-06 | | Release date: | 2024-08-28 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | De novo design of mini-protein binders broadly neutralizing Clostridioides difficile toxin B variants
Nat Commun, 2024
|
|
5WUJ
 
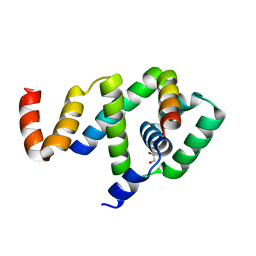 | | Crystal structure of FliF-FliG complex from H. pylori | | Descriptor: | Flagellar M-ring protein, Flagellar motor switch protein FliG, GLYCEROL | | Authors: | Au, S.W, Xue, C, Lam, K.H, Lee, S.H. | | Deposit date: | 2016-12-19 | | Release date: | 2017-12-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the FliF-FliG complex from Helicobacter pylori yields insight into the assembly of the motor MS-C ring in the bacterial flagellum
J. Biol. Chem., 293, 2018
|
|
7CL8
 
 | |
7DRT
 
 | | Human Wntless in complex with Wnt3a | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhong, Q, Zhao, Y, Ye, F, Xiao, Z, Huang, G, Zhang, Y, Lu, P, Xu, W, Zhou, Q, Ma, D. | | Deposit date: | 2020-12-29 | | Release date: | 2021-07-14 | | Last modified: | 2021-09-08 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Cryo-EM structure of human Wntless in complex with Wnt3a.
Nat Commun, 12, 2021
|
|
7D2T
 
 | | Crystal structure of Rsu1/PINCH1_LIM45C complex | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, LIM and senescent cell antigen-like-containing domain protein 1, ... | | Authors: | Yang, H, Wei, Z, Yu, C. | | Deposit date: | 2020-09-17 | | Release date: | 2021-02-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Complex structures of Rsu1 and PINCH1 reveal a regulatory mechanism of the ILK/PINCH/Parvin complex for F-actin dynamics.
Elife, 10, 2021
|
|
7D2S
 
 | | Crystal structure of Rsu1/PINCH1_LIM5C complex | | Descriptor: | GLYCEROL, LIM and senescent cell antigen-like-containing domain protein 1, Ras suppressor protein 1, ... | | Authors: | Yang, H, Wei, Z, Cong, Y. | | Deposit date: | 2020-09-17 | | Release date: | 2021-02-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.653 Å) | | Cite: | Complex structures of Rsu1 and PINCH1 reveal a regulatory mechanism of the ILK/PINCH/Parvin complex for F-actin dynamics.
Elife, 10, 2021
|
|
7D2U
 
 | | Crystal structure of Rsu1/PINCH1_LIM45C complex | | Descriptor: | GLYCEROL, LIM and senescent cell antigen-like-containing domain protein 1, MALONATE ION, ... | | Authors: | Yang, H, Wei, Z, Cong, Y. | | Deposit date: | 2020-09-17 | | Release date: | 2021-02-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Complex structures of Rsu1 and PINCH1 reveal a regulatory mechanism of the ILK/PINCH/Parvin complex for F-actin dynamics.
Elife, 10, 2021
|
|
