1ERO
 
 | | X-RAY CRYSTAL STRUCTURE OF TEM-1 BETA LACTAMASE IN COMPLEX WITH A DESIGNED BORONIC ACID INHIBITOR (1R)-2-PHENYLACETAMIDO-2-(3-CARBOXYPHENYL)ETHYL BORONIC ACID | | Descriptor: | (1R)-2-PHENYLACETAMIDO-2-(3-CARBOXYPHENYL)ETHYL BORONIC ACID, TEM-1 BETA-LACTAMASE | | Authors: | Ness, S, Martin, R, Kindler, A.M, Paetzel, M, Gold, M, Jones, J.B, Strynadka, N.C.J. | | Deposit date: | 2000-04-06 | | Release date: | 2000-05-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-based design guides the improved efficacy of deacylation transition state analogue inhibitors of TEM-1 beta-Lactamase(,).
Biochemistry, 39, 2000
|
|
2JW1
 
 | | Structural characterization of the type III pilotin-secretin interaction in Shigella flexneri by NMR spectroscopy | | Descriptor: | Lipoprotein mxiM, Outer membrane protein mxiD | | Authors: | Okon, M.S, Lario, P.I, Creagh, L, Jung, Y.M.T, Maurelli, A.T, Strynadka, N.C.J, McIntosh, L.P. | | Deposit date: | 2007-10-02 | | Release date: | 2008-09-02 | | Last modified: | 2020-02-19 | | Method: | SOLUTION NMR | | Cite: | Structural Characterization of the Type-III Pilot-Secretin Complex from Shigella flexneri
Structure, 16, 2008
|
|
5HLD
 
 | | E. coli PBP1b in complex with acyl-CENTA and moenomycin | | Descriptor: | (2S)-5-methylidene-2-{(1R)-2-oxo-1-[(thiophen-2-ylacetyl)amino]ethyl}-5,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, MOENOMYCIN, Penicillin-binding protein 1B | | Authors: | King, D.T, Strynadka, N.C.J. | | Deposit date: | 2016-01-14 | | Release date: | 2016-12-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Escherichia coli Penicillin-Binding Protein 1B: Structural Insights into Inhibition.
J. Biol. Chem., 2016
|
|
5HLB
 
 | | E. coli PBP1b in complex with acyl-aztreonam and moenomycin | | Descriptor: | 2-({[(1Z)-1-(2-amino-1,3-thiazol-4-yl)-2-oxo-2-{[(2S,3S)-1-oxo-3-(sulfoamino)butan-2-yl]amino}ethylidene]amino}oxy)-2-methylpropanoic acid, MOENOMYCIN, Penicillin-binding protein 1B | | Authors: | King, D.T, Strynadka, N.C.J. | | Deposit date: | 2016-01-14 | | Release date: | 2016-12-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Escherichia coli Penicillin-Binding Protein 1B: Structural Insights into Inhibition.
J. Biol. Chem., 2016
|
|
1SIW
 
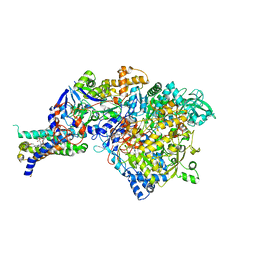 | | Crystal structure of the apomolybdo-NarGHI | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, FE3-S4 CLUSTER, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Rothery, R.A, Bertero, M.G, Cammack, R, Palak, M, Blasco, F, Strynadka, N.C, Weiner, J.H. | | Deposit date: | 2004-03-01 | | Release date: | 2004-06-08 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The catalytic subunit of Escherichia coli nitrate reductase A contains a novel [4Fe-4S] cluster with a high-spin ground state
Biochemistry, 43, 2004
|
|
2FFF
 
 | |
3T05
 
 | |
4G08
 
 | |
4G2S
 
 | |
3T07
 
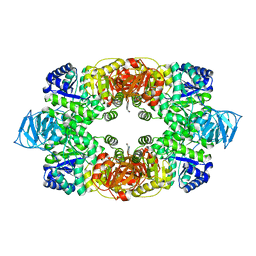 | | Crystal structure of S. aureus Pyruvate Kinase in complex with a naturally occurring bis-indole alkaloid | | Descriptor: | (3S,5R)-3,5-bis(6-bromo-1H-indol-3-yl)piperazin-2-one, PHOSPHATE ION, Pyruvate kinase | | Authors: | Worrall, L.J, Vuckovic, M, Strynadka, N.C.J. | | Deposit date: | 2011-07-19 | | Release date: | 2011-10-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Methicillin-resistant Staphylococcus aureus (MRSA) pyruvate kinase as a target for bis-indole alkaloids with antibacterial activities.
J.Biol.Chem., 286, 2011
|
|
4G1I
 
 | |
4J94
 
 | | Crystal structure of MycP1 from the ESX-1 type VII secretion system | | Descriptor: | Membrane-anchored mycosin mycp1 | | Authors: | Solomonson, M, Wasney, G.A, Watanabe, N, Gruninger, R.J, Prehna, G, Strynadka, N.C.J. | | Deposit date: | 2013-02-15 | | Release date: | 2013-05-01 | | Last modified: | 2013-07-03 | | Method: | X-RAY DIFFRACTION (1.857 Å) | | Cite: | Structure of the Mycosin-1 Protease from the Mycobacterial ESX-1 Protein Type VII Secretion System.
J.Biol.Chem., 288, 2013
|
|
3T0T
 
 | | Crystal structure of S. aureus Pyruvate Kinase | | Descriptor: | N'-[(1E)-1-(1H-benzimidazol-2-yl)ethylidene]-5-bromo-2-hydroxybenzohydrazide, PHOSPHATE ION, Pyruvate kinase | | Authors: | Worrall, L.J, Vuckovic, M, Strynadka, N.C.J. | | Deposit date: | 2011-07-20 | | Release date: | 2012-06-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Cheminformatics-driven discovery of selective, nanomolar inhibitors for staphylococcal pyruvate kinase.
Acs Chem.Biol., 7, 2012
|
|
4X7R
 
 | | Crystal structure of S. aureus TarM G117R mutant in complex with Fondaparinux, alpha-GlcNAc-glycerol and UDP | | Descriptor: | (2S)-2,3-dihydroxypropyl 2-acetamido-2-deoxy-alpha-D-glucopyranoside, 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-3,6-di-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-methyl 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranoside, TarM, ... | | Authors: | Worrall, L.J, Sobhanifar, S, Strynadka, N.C. | | Deposit date: | 2014-12-09 | | Release date: | 2015-02-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure and mechanism of Staphylococcus aureus TarM, the wall teichoic acid alpha-glycosyltransferase.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4X7P
 
 | | Crystal structure of apo S. aureus TarM | | Descriptor: | SULFATE ION, TarM | | Authors: | Worrall, L.J, Sobhanifar, S, Gruninger, R.J, Strynadka, N.C. | | Deposit date: | 2014-12-09 | | Release date: | 2015-02-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure and mechanism of Staphylococcus aureus TarM, the wall teichoic acid alpha-glycosyltransferase.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4X6L
 
 | | Crystal structure of S. aureus TarM in complex with UDP | | Descriptor: | TarM, URIDINE-5'-DIPHOSPHATE | | Authors: | Worrall, L.J, Sobhanifar, S, Gruninger, R.J, Strynadka, N.C. | | Deposit date: | 2014-12-08 | | Release date: | 2015-02-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Structure and mechanism of Staphylococcus aureus TarM, the wall teichoic acid alpha-glycosyltransferase.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
1RO7
 
 | | Structural analysis of the sialyltransferase CstII from Campylobacter jejuni in complex with a substrate analogue, CMP-3FNeuAc. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CYTIDINE-5'-MONOPHOSPHATE-3-FLUORO-N-ACETYL-NEURAMINIC ACID, alpha-2,3/8-sialyltransferase | | Authors: | Chiu, C.P, Watts, A.G, Lairson, L.L, Gilbert, M, Lim, D, Wakarchuk, W.W, Withers, S.G, Strynadka, N.C. | | Deposit date: | 2003-12-01 | | Release date: | 2004-02-03 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of the sialyltransferase CstII from Campylobacter jejuni in complex with a substrate analog.
Nat.Struct.Mol.Biol., 11, 2004
|
|
4X7M
 
 | | Crystal structure of S. aureus TarM G117R mutant in complex with UDP and UDP-GlcNAc | | Descriptor: | TarM, URIDINE-5'-DIPHOSPHATE, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE | | Authors: | Worrall, L.J, Sobhanifar, S, Strynadka, N.C. | | Deposit date: | 2014-12-09 | | Release date: | 2015-03-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and mechanism of Staphylococcus aureus TarM, the wall teichoic acid alpha-glycosyltransferase.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
1OSS
 
 | | T190P STREPTOMYCES GRISEUS TRYPSIN IN COMPLEX WITH BENZAMIDINE | | Descriptor: | BENZAMIDINE, CALCIUM ION, SULFATE ION, ... | | Authors: | Page, M.J, Wong, S.L, Hewitt, J, Strynadka, N.C, MacGillivray, R.T. | | Deposit date: | 2003-03-20 | | Release date: | 2003-08-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Engineering the Primary Substrate Specificity of Streptomyces griseus Trypsin.
Biochemistry, 42, 2003
|
|
1OS8
 
 | | RECOMBINANT STREPTOMYCES GRISEUS TRYPSIN | | Descriptor: | CALCIUM ION, SULFATE ION, trypsin | | Authors: | Page, M.J, Wong, S.L, Hewitt, J, Strynadka, N.C, MacGillivray, R.T. | | Deposit date: | 2003-03-18 | | Release date: | 2003-08-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Engineering the Primary Substrate Specificity of Streptomyces griseus Trypsin.
Biochemistry, 42, 2003
|
|
1ERM
 
 | | X-RAY CRYSTAL STRUCTURE OF TEM-1 BETA LACTAMASE IN COMPLEX WITH A DESIGNED BORONIC ACID INHIBITOR (1R)-1-ACETAMIDO-2-(3-CARBOXYPHENYL)ETHANE BORONIC ACID | | Descriptor: | 1(R)-1-ACETAMIDO-2-(3-CARBOXYPHENYL)ETHYL BORONIC ACID, TEM-1 BETA-LACTAMASE | | Authors: | Ness, S, Martin, R, Kindler, A.M, Paetzel, M, Gold, M, Jones, J.B, Strynadka, N.C.J. | | Deposit date: | 2000-04-06 | | Release date: | 2000-05-10 | | Last modified: | 2012-06-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-based design guides the improved efficacy of deacylation transition state analogue inhibitors of TEM-1 beta-Lactamase(,).
Biochemistry, 39, 2000
|
|
4XP8
 
 | |
4WJ2
 
 | | Mycobacterial protein | | Descriptor: | Antigen MTB48 | | Authors: | Solomonson, M, Strynadka, N.C.J. | | Deposit date: | 2014-09-29 | | Release date: | 2015-02-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of EspB from the ESX-1 type VII secretion system and insights into its export mechanism.
Structure, 23, 2015
|
|
4WJ1
 
 | |
3VZL
 
 | | Crystal structure of the Bacillus circulans endo-beta-(1,4)-xylanase (BcX) N35H mutant | | Descriptor: | Endo-1,4-beta-xylanase, SULFATE ION | | Authors: | Ludwiczek, M.L, D'Angelo, I, Yalloway, G.N, Okon, M, Nielsen, J.E, Strynadka, N.C, Withers, S.G, McIntosh, L.P. | | Deposit date: | 2012-10-15 | | Release date: | 2013-05-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Strategies for modulating the pH-dependent activity of a family 11 glycoside hydrolase
Biochemistry, 52, 2013
|
|
