2NMN
 
 | |
2NN8
 
 | | Crystal structure of human galectin-3 carbohydrate-recognition domain with lactose bound, at 1.35 angstrom resolution | | Descriptor: | CHLORIDE ION, GLYCEROL, Galectin-3, ... | | Authors: | Blanchard, H, Collins, P.M. | | Deposit date: | 2006-10-24 | | Release date: | 2007-03-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Slow diffusion of lactose out of galectin-3 crystals monitored by X-ray crystallography: possible implications for ligand-exchange protocols.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2P3I
 
 | | Crystal structure of Rhesus Rotavirus VP8* at 295K | | Descriptor: | 2-O-methyl-5-N-acetyl-alpha-D-neuraminic acid, SULFATE ION, VP4 | | Authors: | Blanchard, H. | | Deposit date: | 2007-03-09 | | Release date: | 2008-03-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Effects on sialic acid recognition of amino acid mutations in the carbohydrate-binding cleft of the rotavirus spike protein
Glycobiology, 19, 2009
|
|
2P3K
 
 | | Crystal structure of Rhesus rotavirus VP8* at 100K | | Descriptor: | 2-O-methyl-5-N-acetyl-alpha-D-neuraminic acid, GLYCEROL, SULFATE ION, ... | | Authors: | Blanchard, H. | | Deposit date: | 2007-03-09 | | Release date: | 2008-03-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Effects on sialic acid recognition of amino acid mutations in the carbohydrate-binding cleft of the rotavirus spike protein
Glycobiology, 19, 2009
|
|
2P3J
 
 | |
7MYL
 
 | |
7MYM
 
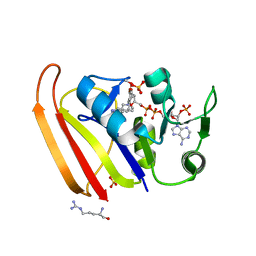 | | Crystal structure of Escherichia coli dihydrofolate reductase in complex with TRIMETHOPRIM and NADPH | | Descriptor: | ARGININE, Dihydrofolate reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Erlandsen, H, Wright, D, Krucinska, J. | | Deposit date: | 2021-05-21 | | Release date: | 2022-06-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Structure-guided functional studies of plasmid-encoded dihydrofolate reductases reveal a common mechanism of trimethoprim resistance in Gram-negative pathogens.
Commun Biol, 5, 2022
|
|
2YAD
 
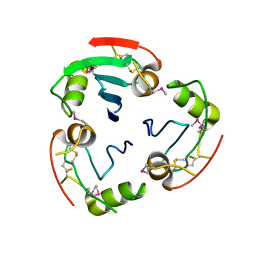 | | BRICHOS domain of Surfactant protein C precursor protein | | Descriptor: | SURFACTANT PROTEIN C BRICHOS DOMAIN | | Authors: | Askarieh, G, Siponen, M.I, Willander, H, Landreh, M, Westermark, P, Nordling, K, Keranen, H, Hermansson, E, Hamvas, A, Nogee, L.M, Bergman, T, Saenz, A, Casals, C, Aqvist, J, Jornvall, H, Presto, J, Johansson, J, Arrowsmith, C.H, Bountra, C, Collins, R, Edwards, A.M, Ekblad, T, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Karlberg, T, Kol, S, Kotenyova, T, Kouznetsova, E, Moche, M, Nyman, T, Nordlund, P, Persson, C, Schuler, H, Thorsell, A.G, Tresaugues, L, van den Berg, S, Wahlberg, E, Weigelt, J, Welin, M, Berglund, H, Knight, S.D. | | Deposit date: | 2011-02-18 | | Release date: | 2012-02-15 | | Last modified: | 2014-08-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | High Resolution Structure of a Bricos Domain and its Implications for Anti-Amyloid Chaperone Activity on Lung Surgactant Protein C.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3T2T
 
 | |
3T1L
 
 | |
3T1M
 
 | |
6BFZ
 
 | | Crystal structure of enolase from E. coli with a mixture of apo form, substrate, and product form | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-PHOSPHOGLYCERIC ACID, Enolase, ... | | Authors: | Erlandsen, H, Wright, D, Krucinska, J. | | Deposit date: | 2017-10-27 | | Release date: | 2018-10-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structural and Functional Studies of Bacterial Enolase, a Potential Target against Gram-Negative Pathogens.
Biochemistry, 58, 2019
|
|
6BFY
 
 | | Crystal structure of enolase from Escherichia coli with bound 2-phosphoglycerate substrate | | Descriptor: | 2-PHOSPHOGLYCERIC ACID, Enolase, GLYCEROL, ... | | Authors: | Erlandsen, H, Wright, D, Krucinska, J. | | Deposit date: | 2017-10-27 | | Release date: | 2018-10-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural and Functional Studies of Bacterial Enolase, a Potential Target against Gram-Negative Pathogens.
Biochemistry, 58, 2019
|
|
2FRX
 
 | | Crystal structure of YebU, a m5C RNA methyltransferase from E.coli | | Descriptor: | Hypothetical protein yebU | | Authors: | Erlandsen, H, Nordlund, P, Hallberg, B.M, Johnson, K.A, Ericsson, U.B. | | Deposit date: | 2006-01-20 | | Release date: | 2006-08-29 | | Last modified: | 2018-05-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The structure of the RNA m5C methyltransferase YebU from Escherichia coli reveals a C-terminal RNA-recruiting PUA domain
J.Mol.Biol., 360, 2006
|
|
3SIT
 
 | |
3SIS
 
 | |
4KAP
 
 | | The Binding of Benzoarylsulfonamide Ligands to Human Carbonic Anhydrase is Insensitive to Formal Fluorination of the Ligand | | Descriptor: | 4,5,6,7-tetrafluoro-1,3-benzothiazole-2-sulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Lange, H, Lockett, M.R, Breiten, B, Heroux, A, Sherman, W, Rappoport, D, Yau, P.O, Snyder, P.W, Whitesides, G.M. | | Deposit date: | 2013-04-22 | | Release date: | 2013-07-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The Binding of Benzoarylsulfonamide Ligands to Human Carbonic Anhydrase is Insensitive to Formal Fluorination of the Ligand.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
1DVI
 
 | | CALPAIN DOMAIN VI WITH CALCIUM BOUND | | Descriptor: | CALCIUM ION, CALPAIN | | Authors: | Cygler, M, Blanchard, H, Grochulski, P. | | Deposit date: | 1997-05-15 | | Release date: | 1998-05-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of a calpain Ca(2+)-binding domain reveals a novel EF-hand and Ca(2+)-induced conformational changes.
Nat.Struct.Biol., 4, 1997
|
|
1PAH
 
 | |
5VWG
 
 | |
6W4Z
 
 | | Galectin-8N terminal domain in complex with Methyl 3-O-[3-O-benzyloxy]-malonyl-beta-D-galactopyranoside | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Galectin-8, ... | | Authors: | Patel, B, Kishor, C, Blanchard, H. | | Deposit date: | 2020-03-12 | | Release date: | 2020-09-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Rational Design and Synthesis of Methyl-beta-d-galactomalonyl Phenyl Esters as Potent Galectin-8 N Antagonists.
J.Med.Chem., 63, 2020
|
|
7RGX
 
 | |
7RGY
 
 | |
7RH0
 
 | |
7RGZ
 
 | |
