5YMT
 
 | |
7KXT
 
 | | Crystal structure of human EED | | Descriptor: | 1-[(4-fluorophenyl)methyl]-N-{1-[2-(4-methoxyphenyl)ethyl]piperidin-4-yl}-1H-benzimidazol-2-amine, Polycomb protein EED, UNKNOWN ATOM OR ION | | Authors: | Zhu, L, Dong, A, Du, D, Liu, Y, Luo, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-04 | | Release date: | 2021-02-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure-Guided Development of Small-Molecule PRC2 Inhibitors Targeting EZH2-EED Interaction.
J.Med.Chem., 64, 2021
|
|
5YMS
 
 | |
5GUT
 
 | | The crystal structure of mouse DNMT1 (731-1602) mutant - N1248A | | Descriptor: | DNA (cytosine-5)-methyltransferase 1, S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION, ... | | Authors: | Chen, S.J, Ye, F. | | Deposit date: | 2016-08-31 | | Release date: | 2017-09-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Biochemical Studies and Molecular Dynamic Simulations Reveal the Molecular Basis of Conformational Changes in DNA Methyltransferase-1.
ACS Chem. Biol., 13, 2018
|
|
5HSI
 
 | | Crystal structure of tyrosine decarboxylase at 1.73 Angstroms resolution | | Descriptor: | MAGNESIUM ION, Putative decarboxylase | | Authors: | Ni, Y, Zhou, J, Zhu, H, Zhang, K. | | Deposit date: | 2016-01-25 | | Release date: | 2016-09-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.732 Å) | | Cite: | Crystal structure of tyrosine decarboxylase and identification of key residues involved in conformational swing and substrate binding
Sci Rep, 6, 2016
|
|
7EVJ
 
 | | Crystal structure of CBP bromodomain liganded with 9c | | Descriptor: | 3-acetyl-1-((3-(1-cyclopropyl-1H-pyrazol-4-yl)-2-fluoro-5-(hydroxymethyl)phenyl)carbamoyl)indolizin-7-yl dimethylcarbamate, CREB-binding protein, GLYCEROL, ... | | Authors: | Xiang, Q, Wang, C, Wu, T, Zhang, Y, Zhang, C, Luo, G, Wu, X, Shen, H, Xu, Y. | | Deposit date: | 2021-05-21 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of 1-(Indolizin-3-yl)ethan-1-ones as CBP Bromodomain Inhibitors for the Treatment of Prostate Cancer.
J.Med.Chem., 65, 2022
|
|
2M30
 
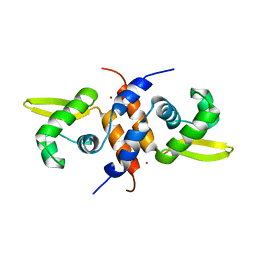 | | Solution NMR refinement of a metal ion bound protein using quantum mechanical/molecular mechanical and molecular dynamics methods | | Descriptor: | Repressor protein, ZINC ION | | Authors: | Chakravorty, D.K, Wang, B.I, Lee, C.I, Guerra, A.J, Giedroc, D.P, Merz Jr, K.M, Arunkumar, A.I, Pennella, M, Kong, X. | | Deposit date: | 2013-01-04 | | Release date: | 2013-05-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR refinement of a metal ion bound protein using metal ion inclusive restrained molecular dynamics methods.
J.Biomol.Nmr, 56, 2013
|
|
5XJR
 
 | |
5XJQ
 
 | |
5XJS
 
 | |
