5L9U
 
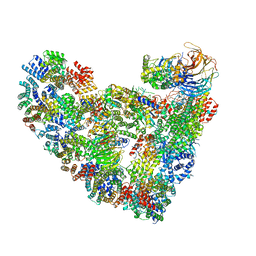 | | Model of human Anaphase-promoting complex/Cyclosome (APC/C-CDH1) with a cross linked Ubiquitin variant-substrate-UBE2C (UBCH10) complex representing key features of multiubiquitination | | Descriptor: | Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 10, Anaphase-promoting complex subunit 11, ... | | Authors: | Brown, N.G, VanderLinden, R, Dube, P, Haselbach, D, Peters, J.M, Stark, H, Schulman, B.A. | | Deposit date: | 2016-06-11 | | Release date: | 2016-09-14 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | Dual RING E3 Architectures Regulate Multiubiquitination and Ubiquitin Chain Elongation by APC/C.
Cell, 165, 2016
|
|
5L9T
 
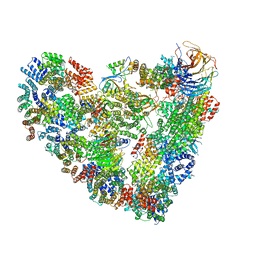 | | Model of human Anaphase-promoting complex/Cyclosome (APC/C-CDH1) with E2 UBE2S poised for polyubiquitination where UBE2S, APC2, and APC11 are modeled into low resolution density | | Descriptor: | Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 10, Anaphase-promoting complex subunit 11, ... | | Authors: | Brown, N.G, VanderLinden, R, Dube, P, Haselbach, D, Peters, J.M, Stark, H, Schulman, B.A. | | Deposit date: | 2016-06-11 | | Release date: | 2016-10-26 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | Dual RING E3 Architectures Regulate Multiubiquitination and Ubiquitin Chain Elongation by APC/C.
Cell, 165, 2016
|
|
8OHD
 
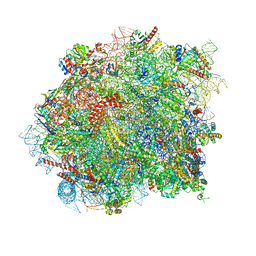 | | 60S ribosomal subunit bound to the E3-UFM1 complex - state 3 (native) | | Descriptor: | 28S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Penchev, I, DaRosa, P.A, Becker, T, Beckmann, R, Kopito, R. | | Deposit date: | 2023-03-21 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | UFM1 E3 ligase promotes recycling of 60S ribosomal subunits from the ER.
Nature, 627, 2024
|
|
6XMT
 
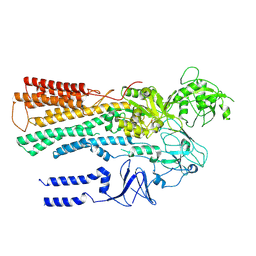 | | Structure of P5A-ATPase Spf1, BeF3-bound form | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, P5A-type ATPase | | Authors: | Park, E, Sim, S.I. | | Deposit date: | 2020-06-30 | | Release date: | 2020-09-23 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The endoplasmic reticulum P5A-ATPase is a transmembrane helix dislocase.
Science, 369, 2020
|
|
6XMQ
 
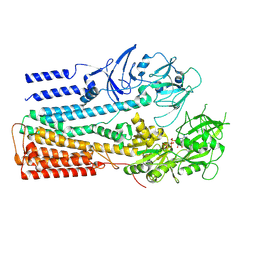 | | Structure of P5A-ATPase Spf1, AMP-PCP-bound form | | Descriptor: | MAGNESIUM ION, P5A-type ATPase, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER | | Authors: | Park, E, Sim, S.I. | | Deposit date: | 2020-06-30 | | Release date: | 2020-09-23 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | The endoplasmic reticulum P5A-ATPase is a transmembrane helix dislocase.
Science, 369, 2020
|
|
6XMU
 
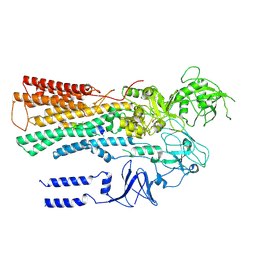 | |
6XMS
 
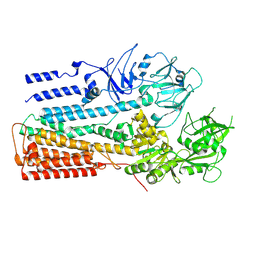 | | Structure of P5A-ATPase Spf1, AlF4-bound form | | Descriptor: | MAGNESIUM ION, P5A-type ATPase, TETRAFLUOROALUMINATE ION | | Authors: | Park, E, Sim, S.I. | | Deposit date: | 2020-06-30 | | Release date: | 2020-09-23 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The endoplasmic reticulum P5A-ATPase is a transmembrane helix dislocase.
Science, 369, 2020
|
|
5C7J
 
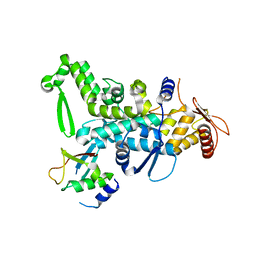 | | CRYSTAL STRUCTURE OF NEDD4 WITH A UB VARIANT | | Descriptor: | E3 ubiquitin-protein ligase NEDD4, Polyubiquitin-C | | Authors: | Walker, J.R, Hu, J, Dong, A, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-06-24 | | Release date: | 2016-03-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | System-Wide Modulation of HECT E3 Ligases with Selective Ubiquitin Variant Probes.
Mol.Cell, 62, 2016
|
|
5C7M
 
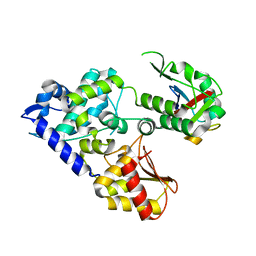 | | CRYSTAL STRUCTURE OF E3 LIGASE ITCH WITH A UB VARIANT | | Descriptor: | E3 ubiquitin-protein ligase Itchy homolog, Polyubiquitin-C | | Authors: | Walker, J.R, Hu, J, Dong, A, Wernimont, A, Zhang, W, Sidhu, S, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-06-24 | | Release date: | 2016-03-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | System-Wide Modulation of HECT E3 Ligases with Selective Ubiquitin Variant Probes.
Mol.Cell, 62, 2016
|
|
8Q7K
 
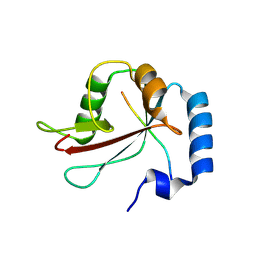 | | IRGQ LIR2 peptide in complex with LC3B | | Descriptor: | 1,2-ETHANEDIOL, Immunity-related GTPase family Q protein, Microtubule-associated proteins 1A/1B light chain 3B | | Authors: | Misra, M, Bailey, H.J, Pomirska, J, Dikic, I. | | Deposit date: | 2023-08-16 | | Release date: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Tumor immune evasion through IRGQ-directed autophagy
To Be Published
|
|
8OJ8
 
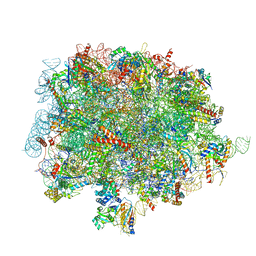 | | 60S ribosomal subunit bound to the E3-UFM1 complex - state 1 (native) | | Descriptor: | 28S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Penchev, I, DaRosa, P.A, Becker, T, Beckmann, R, Kopito, R. | | Deposit date: | 2023-03-24 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | UFM1 E3 ligase promotes recycling of 60S ribosomal subunits from the ER.
Nature, 627, 2024
|
|
8OJ0
 
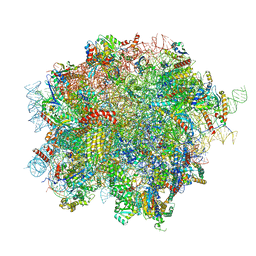 | | 60S ribosomal subunit bound to the E3-UFM1 complex - state 2 (native) | | Descriptor: | 28S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Penchev, I, DaRosa, P.A, Becker, T, Beckmann, R, Kopito, R. | | Deposit date: | 2023-03-23 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | UFM1 E3 ligase promotes recycling of 60S ribosomal subunits from the ER.
Nature, 627, 2024
|
|
8OJ5
 
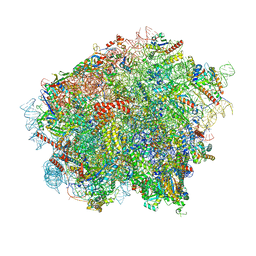 | | 60S ribosomal subunit bound to the E3-UFM1 complex - state 3 (in-vitro reconstitution) | | Descriptor: | 28S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Penchev, I, DaRosa, P.A, Peter, J.J, Kulathu, Y, Becker, T, Beckmann, R, Kopito, R. | | Deposit date: | 2023-03-23 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | UFM1 E3 ligase promotes recycling of 60S ribosomal subunits from the ER.
Nature, 627, 2024
|
|
3HQM
 
 | |
3HU6
 
 | |
3IVQ
 
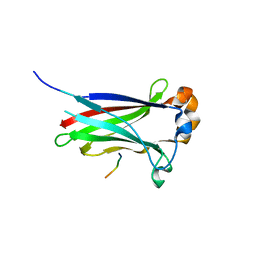 | | Structures of SPOP-Substrate Complexes: Insights into Molecular Architectures of BTB-Cul3 Ubiquitin Ligases: SPOPMATH-CiSBC2 | | Descriptor: | CiSBC2, Speckle-type POZ protein | | Authors: | Schulman, B.A, Miller, D.J, Calabrese, M.F, Seyedin, S. | | Deposit date: | 2009-09-01 | | Release date: | 2009-10-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of SPOP-Substrate Complexes: Insights into Molecular Architectures of BTB-Cul3 Ubiquitin Ligases.
Mol.Cell, 36, 2009
|
|
3IVV
 
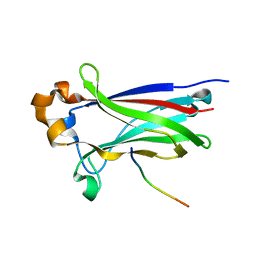 | | Structures of SPOP-Substrate Complexes: Insights into Molecular Architectures of BTB-Cul3 Ubiquitin Ligases: SPOPMATH-PucSBC1_pep1 | | Descriptor: | PucSBC1, Speckle-type POZ protein | | Authors: | Schulman, B.A, Miller, D.J, Calabrese, M.F, Seyedin, S. | | Deposit date: | 2009-09-01 | | Release date: | 2009-10-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structures of SPOP-Substrate Complexes: Insights into Molecular Architectures of BTB-Cul3 Ubiquitin Ligases.
Mol.Cell, 36, 2009
|
|
3HQL
 
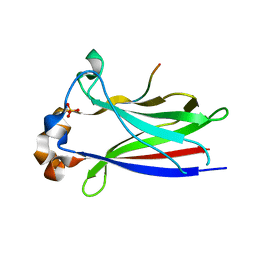 | |
3HVE
 
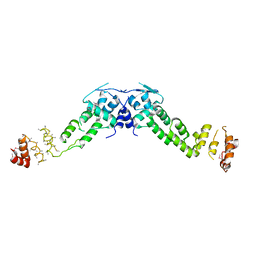 | |
3HQH
 
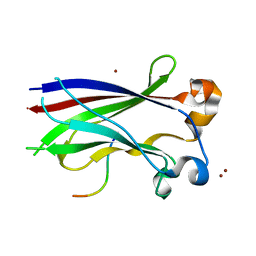 | |
3HTM
 
 | |
3HQI
 
 | |
3HSV
 
 | | Structures of SPOP-Substrate Complexes: Insights into Molecular Architectures of BTB-Cul3 Ubiquitin Ligases: SPOPMATHx-MacroH2ASBCpep2 | | Descriptor: | Core histone macro-H2A.1, SULFATE ION, Speckle-type POZ protein, ... | | Authors: | Zhuang, M, Schulman, B.A, Miller, D. | | Deposit date: | 2009-06-10 | | Release date: | 2009-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Structures of SPOP-substrate complexes: insights into molecular architectures of BTB-Cul3 ubiquitin ligases.
Mol.Cell, 36, 2009
|
|
3IVB
 
 | | Structures of SPOP-Substrate Complexes: Insights into Architectures of BTB-Cul3 Ubiquitin Ligases: SPOPMATH-MacroH2ASBCpep1 | | Descriptor: | CHLORIDE ION, Core histone macro-H2A.1, Speckle-type POZ protein, ... | | Authors: | Schulman, B.A, Miller, D.J, Calabrese, M.F, Seyedin, S. | | Deposit date: | 2009-08-31 | | Release date: | 2009-10-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure 9
To be Published
|
|
5HPS
 
 | |
