2PR9
 
 | | Mu2 adaptin subunit (AP50) of AP2 adaptor (second domain), complexed with GABAA receptor-gamma2 subunit-derived internalization peptide DEEYGYECL | | Descriptor: | AP-2 complex subunit mu-1, GABA(A) receptor subunit gamma-2 peptide | | Authors: | Vahedi-Faridi, A, Haucke, V, Kittler, J.T, Kukhtina, V, Moss, S.J, Saenger, W, Chen, G.-J, Tretter, V, Smith, K, Yan, Z, McAinsh, K, Arancibia-Carcamo, L. | | Deposit date: | 2007-05-04 | | Release date: | 2008-03-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Regulation of synaptic inhibition by phospho-dependent binding of the AP2 complex to a YECL motif in the GABAA receptor gamma2 subunit.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
6JQB
 
 | | The structure of maltooligosaccharide-forming amylase from Pseudomonas saccharophila STB07 with pseudo-maltoheptaose | | Descriptor: | 1,2-ETHANEDIOL, ACARBOSE DERIVED HEPTASACCHARIDE, CALCIUM ION, ... | | Authors: | Li, Z.F, Ban, X.F, Zhang, Z.Q, Li, C.M, Gu, Z.B, Jin, T.C, Li, Y.L, Shang, Y.H. | | Deposit date: | 2019-03-30 | | Release date: | 2020-04-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.101 Å) | | Cite: | Structure of maltotetraose-forming amylase from Pseudomonas saccharophila STB07 provides insights into its product specificity.
Int.J.Biol.Macromol., 154, 2020
|
|
6LKA
 
 | | Crystal Structure of EV71-3C protease with a Novel Macrocyclic Compounds | | Descriptor: | 3C proteinase, ~{N}-[(2~{S})-1-[[(2~{S},3~{S},6~{S},7~{Z},12~{E})-4,9-bis(oxidanylidene)-6-[[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]methyl]-2-phenyl-1,10-dioxa-5-azacyclopentadeca-7,12-dien-3-yl]amino]-3-methyl-1-oxidanylidene-butan-2-yl]-5-methyl-1,2-oxazole-3-carboxamide | | Authors: | Li, P, Wu, S.Q, Xiao, T.Y.C, Li, Y.L, Su, Z.M, Hao, F, Hu, G.P, Hu, J, Lin, F.S, Chen, X.S, Gu, Z.X, He, H.Y, Li, J, Chen, S.H. | | Deposit date: | 2019-12-18 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.033 Å) | | Cite: | Design, synthesis, and evaluation of a novel macrocyclic anti-EV71 agent.
Bioorg.Med.Chem., 28, 2020
|
|
5ZF2
 
 | | Crystal structure of Trxlp from Edwardsiella tarda EIB202 | | Descriptor: | SULFATE ION, Thioredoxin (H-type,TRX-H) | | Authors: | Yang, C, Quan, S. | | Deposit date: | 2018-03-02 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | The Edwardsiella piscicida thioredoxin-like protein inhibits ASK1-MAPKs signaling cascades to promote pathogenesis during infection.
Plos Pathog., 15, 2019
|
|
7MBI
 
 | | Structure of SARS-CoV2 3CL protease covalently bound to peptidomimetic inhibitor | | Descriptor: | 2,4,6-trimethylpyridine-3-carboxylic acid, 3C-like proteinase, 4-methoxy-N-[(2S)-4-methyl-1-oxo-1-({(2S)-3-oxo-1-[(3S)-2-oxopiperidin-3-yl]butan-2-yl}amino)pentan-2-yl]-1H-indole-2-carboxamide | | Authors: | Khan, M.B, Lu, J, Young, H.S, Lemieux, M.J. | | Deposit date: | 2021-03-31 | | Release date: | 2021-07-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Peptidomimetic alpha-Acyloxymethylketone Warheads with Six-Membered Lactam P1 Glutamine Mimic: SARS-CoV-2 3CL Protease Inhibition, Coronavirus Antiviral Activity, and in Vitro Biological Stability.
J.Med.Chem., 65, 2022
|
|
7EXS
 
 | | Thermomicrobium roseum sarcosine oxidase mutant - S320R | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Xin, Y, Shen, C, Tang, M.W, Shi, Y, Guo, Z.T, Gu, Z.H, Shao, J, Zhang, L. | | Deposit date: | 2021-05-28 | | Release date: | 2021-07-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Recreating the natural evolutionary trend in key microdomains provides an effective strategy for engineering of a thermomicrobial N-demethylase.
J.Biol.Chem., 298, 2022
|
|
3TJM
 
 | | Crystal Structure of the Human Fatty Acid Synthase Thioesterase Domain with an Activate Site-Specific Polyunsaturated Fatty Acyl Adduct | | Descriptor: | Fatty acid synthase, methyl (R)-(6Z,9Z,12Z)-octadeca-6,9,12-trien-1-ylphosphonofluoridate | | Authors: | Zhang, W, Zheng, F, Florante, A.Q. | | Deposit date: | 2011-08-24 | | Release date: | 2011-09-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystal structure of FAS thioesterase domain with polyunsaturated fatty acyl adduct and inhibition by dihomo-gamma-linolenic acid.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
8H02
 
 | |
8X01
 
 | |
8YXO
 
 | |
8YXP
 
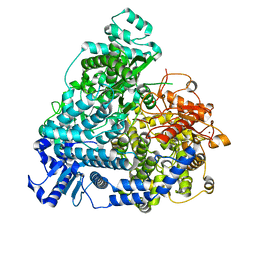 | | Structure of mumps virus L protein (state2) | | Descriptor: | RNA-directed RNA polymerase L, ZINC ION | | Authors: | Li, T.H, Shen, Q.T. | | Deposit date: | 2024-04-02 | | Release date: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structures of the mumps virus polymerase complex via cryo-electron microscopy.
Nat Commun, 15, 2024
|
|
8YXR
 
 | |
8YXM
 
 | |
8YXL
 
 | |
8IYD
 
 | | Tail cap of phage lambda tail | | Descriptor: | Tail tube protein, Tail tube terminator protein | | Authors: | Wang, J.W, Wang, C. | | Deposit date: | 2023-04-04 | | Release date: | 2023-10-18 | | Last modified: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Architecture of the bacteriophage lambda tail.
Structure, 32, 2024
|
|
8IYK
 
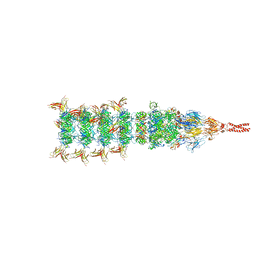 | | Tail tip conformation 1 of phage lambda tail | | Descriptor: | IRON/SULFUR CLUSTER, Tail tip assembly protein I, Tail tip protein L, ... | | Authors: | Wang, J.W, Wang, C. | | Deposit date: | 2023-04-05 | | Release date: | 2023-10-18 | | Last modified: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Architecture of the bacteriophage lambda tail.
Structure, 32, 2024
|
|
8IYL
 
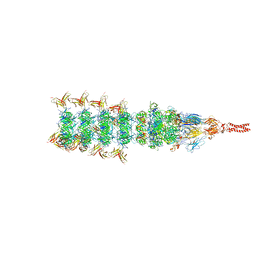 | | Tail tip conformation 2 of phage lambda tail | | Descriptor: | IRON/SULFUR CLUSTER, Tail tip assembly protein I, Tail tip protein L, ... | | Authors: | Wang, J.W, Wang, C. | | Deposit date: | 2023-04-05 | | Release date: | 2023-10-18 | | Last modified: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Architecture of the bacteriophage lambda tail.
Structure, 32, 2024
|
|
8IZL
 
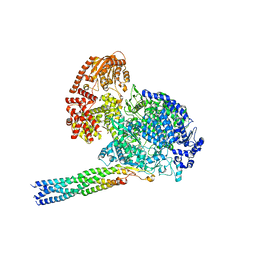 | |
8JVM
 
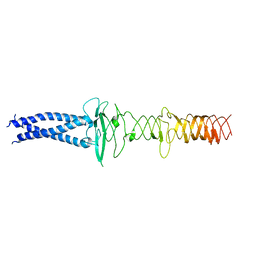 | | AHS-CSF domains of phage lambda tail | | Descriptor: | Tip attachment protein J | | Authors: | Wang, J. | | Deposit date: | 2023-06-28 | | Release date: | 2023-10-18 | | Last modified: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.86 Å) | | Cite: | Architecture of the bacteriophage lambda tail.
Structure, 32, 2024
|
|
8KGE
 
 | |
8KCB
 
 | |
8KCC
 
 | |
7CBQ
 
 | | Crystal structure of PDE4D catalytic domain in complex with Apremilast | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, N-{2-[(1S)-1-(3-ethoxy-4-methoxyphenyl)-2-(methylsulfonyl)ethyl]-1,3-dioxo-2,3-dihydro-1H-isoindol-4-yl}acetamide, ... | | Authors: | Zhang, X.L, Xu, Y.C. | | Deposit date: | 2020-06-13 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Design, synthesis, and biological evaluation of tetrahydroisoquinolines derivatives as novel, selective PDE4 inhibitors for antipsoriasis treatment.
Eur.J.Med.Chem., 211, 2021
|
|
7CBJ
 
 | | Crystal structure of PDE4D catalytic domain in complex with compound 36 | | Descriptor: | (1S)-1-[(7-chloranyl-1H-indol-3-yl)methyl]-6,7-dimethoxy-3,4-dihydro-1H-isoquinoline-2-carbaldehyde, 1,2-ETHANEDIOL, MAGNESIUM ION, ... | | Authors: | Zhang, X.L, Xu, Y.C. | | Deposit date: | 2020-06-12 | | Release date: | 2021-06-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Design, synthesis, and biological evaluation of tetrahydroisoquinolines derivatives as novel, selective PDE4 inhibitors for antipsoriasis treatment.
Eur.J.Med.Chem., 211, 2021
|
|
7WQV
 
 | | Crystal structure of a neutralizing monoclonal antibody (Ab08) in complex with SARS-CoV-2 receptor-binding domain (RBD) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Ab08, ... | | Authors: | Zha, J, Meng, L, Zhang, X, Li, D. | | Deposit date: | 2022-01-26 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A Spike-destructing human antibody effectively neutralizes Omicron-included SARS-CoV-2 variants with therapeutic efficacy.
Plos Pathog., 19, 2023
|
|
