4K3D
 
 | | Crystal structure of bovine antibody BLV1H12 with ultralong CDR H3 | | Descriptor: | BOVINE ANTIBODY WITH ULTRALONG CDR H3, HEAVY CHAIN, LIGHT CHAIN, ... | | Authors: | Ekiert, D.C, Wang, F, Wilson, I.A. | | Deposit date: | 2013-04-10 | | Release date: | 2013-06-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Reshaping antibody diversity.
Cell(Cambridge,Mass.), 153, 2013
|
|
4K3E
 
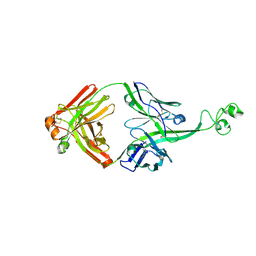 | | Crystal structure of bovine antibody BLV5B8 with ultralong CDR H3 | | Descriptor: | BOVINE ANTIBODY WITH ULTRALONG CDR H3, HEAVY CHAIN, LIGHT CHAIN, ... | | Authors: | Ekiert, D.C, Wang, F, Wilson, I.A. | | Deposit date: | 2013-04-10 | | Release date: | 2013-06-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Reshaping antibody diversity.
Cell(Cambridge,Mass.), 153, 2013
|
|
7MI8
 
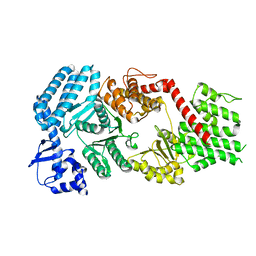 | | Signal subtracted reconstruction of AAA5 and AAA6 domains of dynein in the presence of a pyrazolo-pyrimidinone-based compound, Model 5 | | Descriptor: | Fusion protein of Dynein and Endolysin | | Authors: | Santarossa, C.C, Coudray, N, Urnavicius, L, Ekiert, D.C, Bhabha, G, Kapoor, T.M. | | Deposit date: | 2021-04-16 | | Release date: | 2021-05-26 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Targeting allostery in the Dynein motor domain with small molecule inhibitors.
Cell Chem Biol, 28, 2021
|
|
7MI3
 
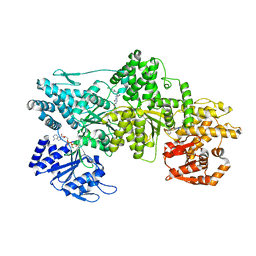 | | Signal subtracted reconstruction of AAA2, AAA3, and AAA4 domains of dynein in the presence of a pyrazolo-pyrimidinone-based compound, Model 4 | | Descriptor: | (8S)-6-(3-bromophenoxy)-2-[1-(4-chlorophenyl)cyclopropyl]-7-hydroxypyrazolo[1,5-a]pyrimidine-3-carbonitrile, ADENOSINE-5'-TRIPHOSPHATE, Fusion protein of Dynein and Endolysin, ... | | Authors: | Santarossa, C.C, Coudray, N, Urnavicius, L, Ekiert, D.C, Bhabha, G, Kapoor, T.M. | | Deposit date: | 2021-04-16 | | Release date: | 2021-05-26 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Targeting allostery in the Dynein motor domain with small molecule inhibitors.
Cell Chem Biol, 28, 2021
|
|
7MI1
 
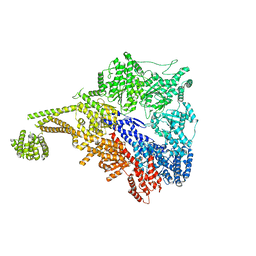 | |
8FEF
 
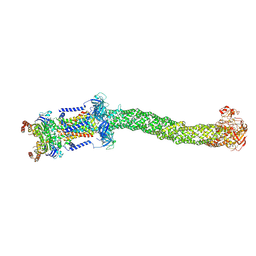 | | Structure of Mce1 transporter from Mycobacterium smegmatis (Map0) | | Descriptor: | ABC transporter, ATP-binding protein,Green fluorescent protein chimera, ABC-transporter integral membrane protein, ... | | Authors: | Chen, J, Bhabha, G, Ekiert, D.C. | | Deposit date: | 2022-12-06 | | Release date: | 2023-02-22 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Structure of an endogenous mycobacterial MCE lipid transporter.
Nature, 620, 2023
|
|
8FED
 
 | | Structure of Mce1-LucB complex from Mycobacterium smegmatis (Map1) | | Descriptor: | ABC transporter, ATP-binding protein,Green fluorescent protein chimera, ABC-transporter integral membrane protein, ... | | Authors: | Chen, J, Bhabha, G, Ekiert, D.C. | | Deposit date: | 2022-12-06 | | Release date: | 2023-02-22 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Structure of an endogenous mycobacterial MCE lipid transporter.
Nature, 620, 2023
|
|
8FEE
 
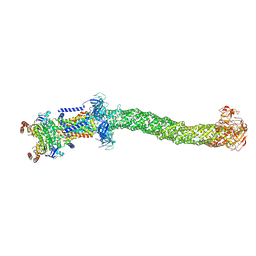 | | Structure of Mce1 transporter from Mycobacterium smegmatis in the absence of LucB (Map2) | | Descriptor: | ABC transporter, ATP-binding protein,Green fluorescent protein chimera, ABC-transporter integral membrane protein, ... | | Authors: | Chen, J, Bhabha, G, Ekiert, D.C. | | Deposit date: | 2022-12-06 | | Release date: | 2023-02-22 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure of an endogenous mycobacterial MCE lipid transporter.
Nature, 620, 2023
|
|
7UDK
 
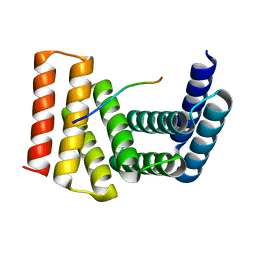 | | Crystal structure of designed helical repeat protein RPB_LRP2_R4 bound to LRPx4 peptide | | Descriptor: | 4xLRP, Designed helical repeat protein (DHR) RPB_LRP2_R4 | | Authors: | Chang, Y, Redler, R.L, Bhabha, G, Ekiert, D.C. | | Deposit date: | 2022-03-20 | | Release date: | 2023-03-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | De novo design of modular peptide-binding proteins by superhelical matching.
Nature, 616, 2023
|
|
7UDL
 
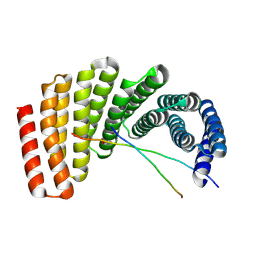 | | Crystal structure of designed helical repeat protein RPB_PLP1_R6 bound to PLPx6 peptide | | Descriptor: | 1,2-ETHANEDIOL, 6xPLP Peptide, Designed helical repeat protein (DHR) RPB_PLP1_R6 | | Authors: | Chang, Y, Redler, R.L, Bhabha, G, Ekiert, D.C. | | Deposit date: | 2022-03-20 | | Release date: | 2023-03-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | De novo design of modular peptide-binding proteins by superhelical matching.
Nature, 616, 2023
|
|
7UDN
 
 | |
7UDM
 
 | | Crystal structure of designed helical repeat protein RPB_PLP1_R6 in alternative conformation 1 (with peptide) | | Descriptor: | 6xPLP, Designed helical repeat protein (DHR) RPB_PLP1_R6 | | Authors: | Chang, Y, Redler, R.L, Bhabha, G, Ekiert, D.C. | | Deposit date: | 2022-03-20 | | Release date: | 2023-03-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | De novo design of modular peptide-binding proteins by superhelical matching.
Nature, 616, 2023
|
|
7UDO
 
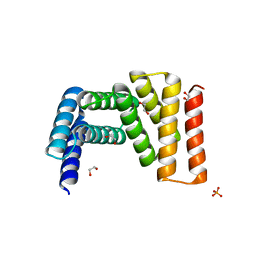 | | Crystal structure of designed helical repeat protein RPB_LRP2_R4 (proteolysis fragment?), forming pseudopolymeric filaments | | Descriptor: | 1,2-ETHANEDIOL, Designed helical repeat protein (DHR) RPB_LRP2_R4, PHOSPHATE ION | | Authors: | Redler, R.L, Chang, Y, Bhabha, G, Ekiert, D.C. | | Deposit date: | 2022-03-20 | | Release date: | 2023-03-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | De novo design of modular peptide-binding proteins by superhelical matching.
Nature, 616, 2023
|
|
7UE2
 
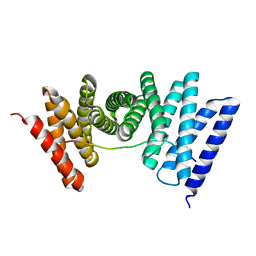 | |
7UPQ
 
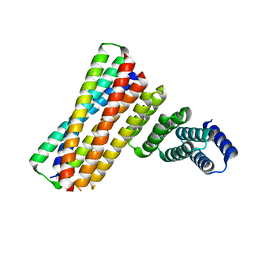 | |
7UPO
 
 | |
7UPP
 
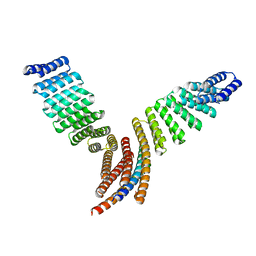 | |
4PEJ
 
 | | Crystal structure of a computationally designed retro-aldolase, RA110.4 (Cys free) | | Descriptor: | Retro-aldolase | | Authors: | Bhabha, G, Zhang, X, Liu, Y, Ekiert, D.C. | | Deposit date: | 2014-04-23 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | De novo-designed enzymes as small-molecule-regulated fluorescence imaging tags and fluorescent reporters.
J.Am.Chem.Soc., 136, 2014
|
|
4PEK
 
 | | Crystal structure of a computationally designed retro-aldolase, RA114.3 | | Descriptor: | Retro-aldolase | | Authors: | Bhabha, G, Zhang, X, Liu, Y, Ekiert, D.C. | | Deposit date: | 2014-04-23 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | De novo-designed enzymes as small-molecule-regulated fluorescence imaging tags and fluorescent reporters.
J.Am.Chem.Soc., 136, 2014
|
|
3QL3
 
 | | Re-refined coordinates for PDB entry 1RX2 | | Descriptor: | Dihydrofolate reductase, FOLIC ACID, MANGANESE (II) ION, ... | | Authors: | Bhabha, G, Ekiert, D.C, Wright, P.E, Wilson, I.A. | | Deposit date: | 2011-02-02 | | Release date: | 2011-04-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A dynamic knockout reveals that conformational fluctuations influence the chemical step of enzyme catalysis.
Science, 332, 2011
|
|
4OU1
 
 | | Crystal structure of a computationally designed retro-aldolase covalently bound to folding probe 1 [(6-methoxynaphthalen-2-yl)(oxiran-2-yl)methanol] | | Descriptor: | (1S,2S)-1-(6-methoxynaphthalen-2-yl)propane-1,2-diol, BENZOIC ACID, PHOSPHATE ION, ... | | Authors: | Bhabha, G, Zhang, X, Ekiert, D.C. | | Deposit date: | 2014-02-14 | | Release date: | 2014-03-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Small molecule probes to quantify the functional fraction of a specific protein in a cell with minimal folding equilibrium shifts.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3QL0
 
 | | Crystal structure of N23PP/S148A mutant of E. coli dihydrofolate reductase | | Descriptor: | Dihydrofolate reductase, FOLIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Bhabha, G, Ekiert, D.C, Wright, P.E, Wilson, I.A. | | Deposit date: | 2011-02-02 | | Release date: | 2011-04-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A dynamic knockout reveals that conformational fluctuations influence the chemical step of enzyme catalysis.
Science, 332, 2011
|
|
6XBD
 
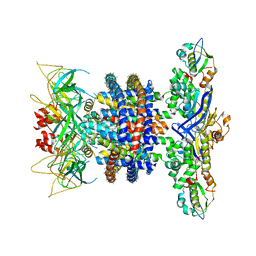 | | Cryo-EM structure of MlaFEDB in nanodiscs with phospholipid substrates | | Descriptor: | DI-PALMITOYL-3-SN-PHOSPHATIDYLETHANOLAMINE, MSP1D1, Phospholipid ABC transporter permease protein MlaE, ... | | Authors: | Coudray, N, Isom, G.L, MacRae, M.R, Saiduddin, M, Ekiert, D.C, Bhabha, G. | | Deposit date: | 2020-06-05 | | Release date: | 2020-07-01 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Structure of bacterial phospholipid transporter MlaFEDB with substrate bound.
Elife, 9, 2020
|
|
6XGZ
 
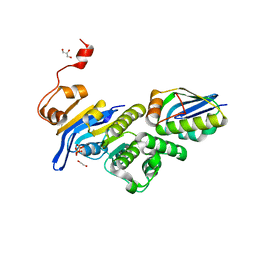 | | Crystal structure of E. coli MlaFB ABC transport subunits in the monomeric state | | Descriptor: | 1,2-ETHANEDIOL, ABC transporter maintaining OM lipid asymmetry, cytoplasmic STAS component, ... | | Authors: | Chang, Y, Bhabha, G, Ekiert, D.C. | | Deposit date: | 2020-06-18 | | Release date: | 2020-07-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of MlaFB uncovers novel mechanisms of ABC transporter regulation.
Elife, 9, 2020
|
|
6XGY
 
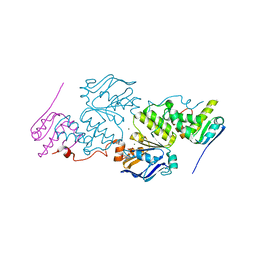 | | Crystal structure of E. coli MlaFB ABC transport subunits in the dimeric state | | Descriptor: | ABC transporter maintaining OM lipid asymmetry, cytoplasmic STAS component, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Chang, Y, Bhabha, G, Ekiert, D.C. | | Deposit date: | 2020-06-18 | | Release date: | 2020-07-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of MlaFB uncovers novel mechanisms of ABC transporter regulation.
Elife, 9, 2020
|
|
