1EA4
 
 | | TRANSCRIPTIONAL REPRESSOR COPG/22bp dsDNA COMPLEX | | Descriptor: | DNA (5'-D(*TP*AP*AP*CP*CP*GP*TP*GP *CP*AP*CP*TP*CP*AP*AP*TP*GP*CP*AP*AP*TP*C)-3'), DNA(5'-D(*AP*GP*AP*TP*TP*GP*CP*AP*TP *TP*GP*AP*GP*TP*GP*CP*AP*CP*GP*GP*TP*T)-3'), TRANSCRIPTIONAL REPRESSOR COPG | | Authors: | Gomis-Rueth, F.X, Costa, M, Sola, M, Acebo, P, Eritja, R, Espinosa, M, Solar, G.D, Coll, M. | | Deposit date: | 2000-11-05 | | Release date: | 2001-07-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Plasmid Transcriptional Repressor Copg Oligomerises to Render Helical Superstructures Unbound and in Complexes with Oligonucleotides
J.Mol.Biol., 310, 2001
|
|
1NDN
 
 | | MOLECULAR STRUCTURE OF NICKED DNA. MODEL T4 | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*AP*AP*CP*GP*CP*G)-3'), DNA (5'-D(*CP*GP*CP*GP*TP*T)-3'), DNA (5'-D(*TP*TP*CP*GP*CP*G)-3') | | Authors: | Aymani, J, Coll, M, Van Der Marel, G.A, Van Boom, J.H, Wang, A.H.-J, Rich, A. | | Deposit date: | 1992-01-15 | | Release date: | 1992-07-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular structure of nicked DNA: a substrate for DNA repair enzymes.
Proc.Natl.Acad.Sci.USA, 87, 1990
|
|
6EY7
 
 | | Human cytomegalovirus terminase nuclease domain, Mn soaked, inhibitor bound | | Descriptor: | 4-[(4-fluorophenyl)methyl-methyl-amino]-2,4-bis(oxidanylidene)butanoic acid, CHLORIDE ION, MANGANESE (II) ION, ... | | Authors: | Bongarzone, S, Nadal, M, Kaczmarska, Z, Machon, C, Alvarez, M, Albericio, F, Coll, M. | | Deposit date: | 2017-11-10 | | Release date: | 2018-10-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure-Driven Discovery of alpha , gamma-Diketoacid Inhibitors Against UL89 Herpesvirus Terminase.
Acs Omega, 3, 2018
|
|
3DKX
 
 | | Crystal Structure of the replication initiator protein encoded on plasmid pMV158 (RepB), trigonal form, to 2.7 Ang resolution | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Boer, D.R, Ruiz-Maso, J.A, Blanco, A.G, Vives-Llacer, M, Uson, I, Gomis-Ruth, F.X, Espinosa, M, Del Solar, G, Coll, M. | | Deposit date: | 2008-06-26 | | Release date: | 2009-06-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Plasmid replication initiator RepB forms a hexamer reminiscent of ring helicases and has mobile nuclease domains
Embo J., 28, 2009
|
|
3DKY
 
 | | Crystal Structure of the replication initiator protein encoded on plasmid pMV158 (RepB), tetragonal form, to 3.6 Ang resolution | | Descriptor: | MANGANESE (II) ION, Replication protein repB | | Authors: | Boer, D.R, Ruiz-Maso, J.A, Blanco, A.G, Vives-Llacer, M, Uson, I, Gomis-Ruth, F.X, Espinosa, M, Del Solar, G, Coll, M. | | Deposit date: | 2008-06-26 | | Release date: | 2009-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Plasmid replication initiator RepB forms a hexamer reminiscent of ring helicases and has mobile nuclease domains
Embo J., 28, 2009
|
|
7ZQV
 
 | | Structure of the SARS-CoV-2 main protease in complex with AG7404 | | Descriptor: | 3C-like proteinase nsp5, ethyl (4R)-4-({(2S)-2-[3-{[(5-methyl-1,2-oxazol-3-yl)carbonyl]amino}-2-oxopyridin-1(2H)-yl]pent-4-ynoyl}amino)-5-[(3S)-2-oxopyrrolidin-3-yl]pentanoate | | Authors: | Fabrega-Ferrer, M, Herrera-Morande, A, Perez-Saavedra, J, Coll, M. | | Deposit date: | 2022-05-03 | | Release date: | 2022-12-28 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structure and inhibition of SARS-CoV-1 and SARS-CoV-2 main proteases by oral antiviral compound AG7404.
Antiviral Res., 208, 2022
|
|
7ZQW
 
 | | Structure of the SARS-CoV-1 main protease in complex with AG7404 | | Descriptor: | 3C-like proteinase nsp5, ethyl (4R)-4-({(2S)-2-[3-{[(5-methyl-1,2-oxazol-3-yl)carbonyl]amino}-2-oxopyridin-1(2H)-yl]pent-4-ynoyl}amino)-5-[(3S)-2-oxopyrrolidin-3-yl]pentanoate | | Authors: | Muriel-Goni, S, Fabrega-Ferrer, M, Herrera-Morande, A, Coll, M. | | Deposit date: | 2022-05-03 | | Release date: | 2022-12-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structure and inhibition of SARS-CoV-1 and SARS-CoV-2 main proteases by oral antiviral compound AG7404.
Antiviral Res., 208, 2022
|
|
1BAY
 
 | | GLUTATHIONE S-TRANSFERASE YFYF CYS 47-CARBOXYMETHYLATED CLASS PI, FREE ENZYME | | Descriptor: | GLUTATHIONE S-TRANSFERASE CLASS PI | | Authors: | Vega, M.C, Coll, M. | | Deposit date: | 1996-11-02 | | Release date: | 1997-11-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The three-dimensional structure of Cys-47-modified mouse liver glutathione S-transferase P1-1. Carboxymethylation dramatically decreases the affinity for glutathione and is associated with a loss of electron density in the alphaB-310B region.
J.Biol.Chem., 273, 1998
|
|
1OMH
 
 | | Conjugative Relaxase TrwC in complex with OriT Dna. Metal-free structure. | | Descriptor: | DNA OLIGONUCLEOTIDE, SULFATE ION, trwC protein | | Authors: | Guasch, A, Lucas, M, Moncalian, G, Cabezas, M, Perez-Luque, R, Gomis-Ruth, F.X, de la Cruz, F, Coll, M. | | Deposit date: | 2003-02-25 | | Release date: | 2003-11-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Recognition and processing of the origin of transfer DNA by conjugative relaxase TrwC.
Nat.Struct.Biol., 10, 2003
|
|
1OSB
 
 | | Conjugative Relaxase TrwC in complex with OriT Dna. Metal-free structure. | | Descriptor: | Dna oligonucleotide, SULFATE ION, TrwC protein | | Authors: | Guasch, A, Lucas, M, Moncalian, G, Cabezas, M, Perez-Luque, R, Gomis-Ruth, F.X, de la Cruz, F, Coll, M. | | Deposit date: | 2003-03-19 | | Release date: | 2003-11-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Recognition and processing of the origin of transfer DNA by conjugative relaxase TrwC.
Nat.Struct.Biol., 10, 2003
|
|
4GQJ
 
 | | Complex of a binuclear Ruthenium compound D,D-([mu-(11,11')-bi(dppz)-(1,10-phenanthroline)4-Ru2]4+) bound to d(CGTACG) | | Descriptor: | (mu-11,11'-bidipyrido[3,2-a:2',3'-c]phenazine-1kappa~2~N~4~,N~5~:2kappa~2~N~4'~,N~5'~)[tetrakis(1,10-phenanthroline-kappa~2~N~1~,N~10~)]diruthenium, DNA (5'-D(*CP*GP*TP*AP*CP*G)-3') | | Authors: | Boer, D.R, Coll, M. | | Deposit date: | 2012-08-23 | | Release date: | 2014-03-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.951 Å) | | Cite: | Thread Insertion of a Bis(dipyridophenazine) Diruthenium Complex into the DNA Double Helix by the Extrusion of AT Base Pairs and Cross-Linking of DNA Duplexes.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
8AMT
 
 | | OBD-RepB pMV158 domain | | Descriptor: | CHLORIDE ION, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Machon, C, Amodio, J, Boer, R.D, Ruiz-Maso, J.A, del Solar, G, Coll, M. | | Deposit date: | 2022-08-04 | | Release date: | 2023-02-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structures of pMV158 replication initiator RepB with and without DNA reveal a flexible dual-function protein.
Nucleic Acids Res., 51, 2023
|
|
8AMU
 
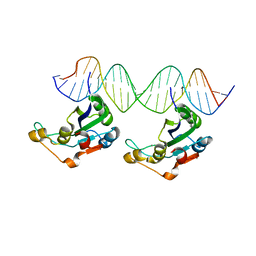 | | RepB pMV158 OBD domain bound to DDR region | | Descriptor: | DNA (5'-D(*AP*AP*AP*AP*GP*TP*CP*GP*CP*CP*GP*AP*AP*AP*AP*GP*TP*CP*GP*CP*CP*GP*A)-3'), DNA (5'-D(*TP*CP*GP*GP*CP*GP*AP*CP*TP*TP*TP*TP*CP*GP*GP*CP*GP*AP*CP*TP*TP*TP*T)-3'), MANGANESE (II) ION, ... | | Authors: | Amodio, J, Machon, C, Boer, R.D, Ruiz-Maso, J.A, del Solar, G, Coll, M. | | Deposit date: | 2022-08-04 | | Release date: | 2023-02-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of pMV158 replication initiator RepB with and without DNA reveal a flexible dual-function protein.
Nucleic Acids Res., 51, 2023
|
|
8AMV
 
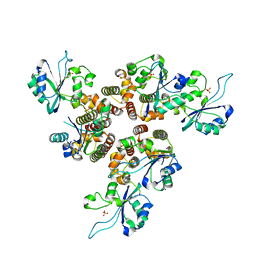 | | RepB pMV158 hexamer | | Descriptor: | PHOSPHATE ION, Replication protein RepB, SODIUM ION | | Authors: | Machon, C, Amodio, J, Boer, R.D, Ruiz-Maso, J.A, del Solar, G, Coll, M. | | Deposit date: | 2022-08-04 | | Release date: | 2023-02-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Structures of pMV158 replication initiator RepB with and without DNA reveal a flexible dual-function protein.
Nucleic Acids Res., 51, 2023
|
|
3I1D
 
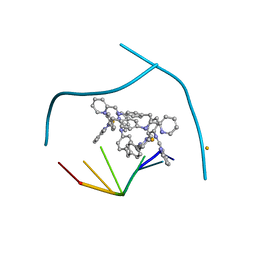 | | Distinct recognition of three-way DNA junctions by the two enantiomers of a metallo-supramolecular cylinder ('helicate') | | Descriptor: | 5'-D(*CP*GP*TP*AP*CP*G)-3', FE (II) ION, N-[(1E)-PYRIDIN-2-YLMETHYLENE]-N-[4-(4-{[(1E)-PYRIDIN-2-YLMETHYLENE]AMINO}BENZYL)PHENYL]AMINE | | Authors: | Boer, D.R, Uson, I, Hannon, M.J, Coll, M. | | Deposit date: | 2009-06-26 | | Release date: | 2011-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Self-Assembly of Functionalizable Two-Component 3D DNA Arrays through the Induced Formation of DNA Three-Way-Junction Branch Points by Supramolecular Cylinders.
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
3TQ6
 
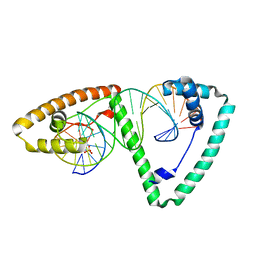 | | Crystal structure of human mitochondrial transcription factor A, TFAM or mtTFA, bound to the light strand promoter LSP | | Descriptor: | DI(HYDROXYETHYL)ETHER, DNA (5'-D(*GP*TP*TP*AP*GP*TP*TP*GP*GP*GP*GP*GP*GP*TP*GP*AP*CP*TP*GP*TP*TP*A)-3'), DNA (5'-D(*TP*AP*AP*CP*AP*GP*TP*CP*AP*CP*CP*CP*CP*CP*CP*AP*AP*CP*(BRU)P*AP*AP*C)-3'), ... | | Authors: | Rubio-Cosials, A, Sydow, J.F, Jimenez-Menendez, N, Fernandez-Millan, P, Montoya, J, Jacobs, H.T, Coll, M, Bernado, P, Sola, M. | | Deposit date: | 2011-09-09 | | Release date: | 2011-11-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Human mitochondrial transcription factor A induces a U-turn structure in the light strand promoter.
Nat.Struct.Mol.Biol., 18, 2011
|
|
3T72
 
 | | PhoB(E)-Sigma70(4)-(RNAP-Betha-flap-tip-helix)-DNA Transcription Activation Sub-Complex | | Descriptor: | PHO BOX DNA (STRAND 1), PHO BOX DNA (STRAND 2), Phosphate regulon transcriptional regulatory protein phoB, ... | | Authors: | Blanco, A.G, Canals, A, Bernues, J, Sola, M, Coll, M. | | Deposit date: | 2011-07-29 | | Release date: | 2011-09-21 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (4.33 Å) | | Cite: | The structure of a transcription activation subcomplex reveals how sigma (70) is recruited to PhoB promoters.
Embo J., 30, 2011
|
|
4LVI
 
 | | MobM Relaxase Domain (MOBV; Mob_Pre) bound to plasmid pMV158 oriT DNA (22nt). Mn-bound crystal structure at pH 4.6 | | Descriptor: | ACTTTAT oligonucleotide, ATAAAGTATAGTGTG oligonucleotide, GLYCEROL, ... | | Authors: | Pluta, R, Boer, D.R, Coll, M. | | Deposit date: | 2013-07-26 | | Release date: | 2014-09-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of a histidine-DNA nicking/joining mechanism for gene transfer and promiscuous spread of antibiotic resistance.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4LVL
 
 | | MobM Relaxase Domain (MOBV; Mob_Pre) bound to plasmid pMV158 oriT DNA (22nt+3'Thiophosphate). Mn-bound crystal structure at pH 6.8 | | Descriptor: | CHLORIDE ION, DNA (5'-D(*AP*CP*TP*TP*TP*AP*T)-3'), DNA (5'-D(*AP*TP*AP*AP*AP*GP*TP*AP*TP*AP*GP*TP*GP*TP*GP*(TS6))-3'), ... | | Authors: | Pluta, R, Boer, D.R, Coll, M. | | Deposit date: | 2013-07-26 | | Release date: | 2014-09-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of a histidine-DNA nicking/joining mechanism for gene transfer and promiscuous spread of antibiotic resistance.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4LVM
 
 | | MobM Relaxase Domain (MOBV; Mob_Pre) bound to plasmid pMV158 oriT DNA (23nt). Mn-bound crystal structure at pH 6.5 | | Descriptor: | ACTTTAT oligonucleotide, ATAAAGTATAGTGTGT oligonucleotide, CHLORIDE ION, ... | | Authors: | Pluta, R, Boer, D.R, Coll, M. | | Deposit date: | 2013-07-26 | | Release date: | 2014-09-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis of a histidine-DNA nicking/joining mechanism for gene transfer and promiscuous spread of antibiotic resistance.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4LVK
 
 | | MobM Relaxase Domain (MOBV; Mob_Pre) bound to plasmid pMV158 oriT DNA (22nt+3'Phosphate). Mn-bound crystal structure at pH 4.6 | | Descriptor: | ACTTTAT oligonucleotide, ATAAAGTATAGTGTGpo oligonucleotide, MANGANESE (II) ION, ... | | Authors: | Pluta, R, Boer, D.R, Coll, M. | | Deposit date: | 2013-07-26 | | Release date: | 2014-09-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structural basis of a histidine-DNA nicking/joining mechanism for gene transfer and promiscuous spread of antibiotic resistance.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4LVJ
 
 | | MobM Relaxase Domain (MOBV; Mob_Pre) bound to plasmid pMV158 oriT DNA (22nt). Mn-bound crystal structure at pH 5.5 | | Descriptor: | ACETATE ION, ACTTTAT oligonucleotide, ATAAAGTATAGTGTG oligonucleotide, ... | | Authors: | Pluta, R, Boer, D.R, Coll, M. | | Deposit date: | 2013-07-26 | | Release date: | 2014-09-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structural basis of a histidine-DNA nicking/joining mechanism for gene transfer and promiscuous spread of antibiotic resistance.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
1GL6
 
 | | Plasmid coupling protein TrwB in complex with the non-hydrolysable GTP analogue GDPNP | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ATPASE, CHLORIDE ION, ... | | Authors: | Gomis-Ruth, F.X, Moncalian, G, De La cruz, F, Coll, M. | | Deposit date: | 2001-08-28 | | Release date: | 2002-05-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Bacterial Conjugation Protein Trwb Resembles Ring Helicases and F1-ATPase
Nature, 409, 2001
|
|
4MNB
 
 | | Crystal Structure of a complex between the marine anticancer drug Variolin B and DNA | | Descriptor: | 5'-D(*CP*GP*TP*AP*CP*G)-3', 9-amino-5-(2-aminopyrimidin-4-yl)pyrido[3',2':4,5]pyrrolo[1,2-c]pyrimidin-4-ol, COBALT (II) ION, ... | | Authors: | Canals, A, Arribas-Bosacoma, R, Alvarez, M, Albericio, F, Aymami, J, Coll, M. | | Deposit date: | 2013-09-10 | | Release date: | 2015-03-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Intercalative DNA binding of the marine anticancer drug variolin B.
Sci Rep, 7, 2017
|
|
4ICU
 
 | | Ubiquitin-like domain of human tubulin folding cofactor E - crystal from A | | Descriptor: | Tubulin-specific chaperone E | | Authors: | Janowski, R, Boutin, M, Zabala, J.C, Coll, M. | | Deposit date: | 2012-12-11 | | Release date: | 2014-06-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structure of the complex between alpha-tubulin, TBCE and TBCB reveals a tubulin dimer dissociation mechanism.
J.Cell.Sci., 128, 2015
|
|
