1CQP
 
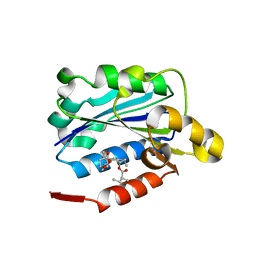 | | CRYSTAL STRUCTURE ANALYSIS OF THE COMPLEX LFA-1 (CD11A) I-DOMAIN / LOVASTATIN AT 2.6 A RESOLUTION | | Descriptor: | ANTIGEN CD11A (P180), LOVASTATIN, MAGNESIUM ION | | Authors: | Kallen, J, Welzenbach, K, Ramage, P, Geyl, D, Kriwacki, R, Legge, G, Cottens, S, Weitz-Schmidt, G, Hommel, U. | | Deposit date: | 1999-08-10 | | Release date: | 2000-08-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for LFA-1 inhibition upon lovastatin binding to the CD11a I-domain.
J.Mol.Biol., 292, 1999
|
|
1BWU
 
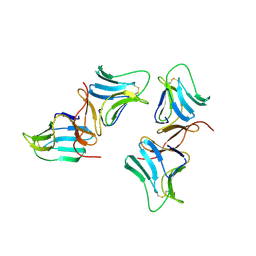 | | MANNOSE-SPECIFIC AGGLUTININ (LECTIN) FROM GARLIC (ALLIUM SATIVUM) BULBS COMPLEXED WITH ALPHA-D-MANNOSE | | Descriptor: | PROTEIN (AGGLUTININ), alpha-D-mannopyranose | | Authors: | Chandra, N.R, Ramachandraiah, G, Bachhawat, K, Dam, T.K, Surolia, A, Vijayan, M. | | Deposit date: | 1998-09-28 | | Release date: | 1999-01-20 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a dimeric mannose-specific agglutinin from garlic: quaternary association and carbohydrate specificity.
J.Mol.Biol., 285, 1999
|
|
1XDG
 
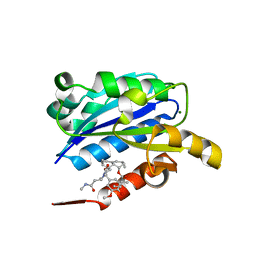 | | X-ray structure of LFA-1 I-domain in complex with LFA878 at 2.1A resolution | | Descriptor: | (1S,3R,8AS)-8-(2-{(4S,6S)-3-(4-HYDROXY-3-METHOXYBENZYL)-4-[2-(METHYLAMINO)-2-OXOETHYL]-2-OXO-1,3-OXAZINAN-6-YL}ETHYL)-3 ,7-DIMETHYL-1,2,3,7,8,8A-HEXAHYDRONAPHTHALEN-1-YL (2R)-2-METHYLBUTANOATE, Integrin alpha-L, MAGNESIUM ION | | Authors: | Weitz-Schmidt, G, Welzenbach, K, Dawson, J, Kallen, J. | | Deposit date: | 2004-09-06 | | Release date: | 2004-09-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Improved lymphocyte function-associated antigen-1 (LFA-1) inhibition by statin derivatives: molecular basis determined by x-ray analysis and monitoring of LFA-1 conformational changes in vitro and ex vivo
J.Biol.Chem., 279, 2004
|
|
1XDD
 
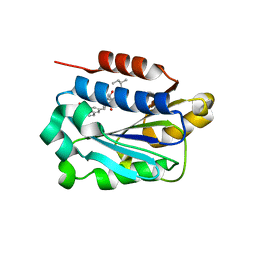 | | X-ray structure of LFA-1 I-domain in complex with LFA703 at 2.2A resolution | | Descriptor: | 8-[2-((2S)-4-HYDROXY-1-{[5-(HYDROXYMETHYL)-6-METHOXY-2-NAPHTHYL]METHYL}-6-OXOPIPERIDIN-2-YL)ETHYL]-3,7-DIMETHYL-1,2,3,7,8,8A-HEXAHYDRONAPHTHALEN-1-YL 2-METHYLBUTANOATE, Integrin alpha-L, MAGNESIUM ION | | Authors: | Weitz-Schmidt, G, Welzenbach, K, Dawson, J, Kallen, J. | | Deposit date: | 2004-09-06 | | Release date: | 2004-09-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Improved lymphocyte function-associated antigen-1 (LFA-1) inhibition by statin derivatives: molecular basis determined by x-ray analysis and monitoring of LFA-1 conformational changes in vitro and ex vivo
J.Biol.Chem., 279, 2004
|
|
1XUO
 
 | | X-ray structure of LFA-1 I-domain bound to a 1,4-diazepane-2,5-dione inhibitor at 1.8A resolution | | Descriptor: | (2R)-2-[3-ISOBUTYL-2,5-DIOXO-4-(QUINOLIN-3-YLMETHYL)-1,4-DIAZEPAN-1-YL]-N-METHYL-3-(2-NAPHTHYL)PROPANAMIDE, Integrin alpha-L, MAGNESIUM ION | | Authors: | Wattanasin, S, Kallen, J, Myers, S, Guo, Q, Sabio, M, Ehrhardt, C, Albert, R, Hommel, U, Weckbecker, G, Welzenbach, K. | | Deposit date: | 2004-10-26 | | Release date: | 2005-10-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1,4-Diazepane-2,5-diones as novel inhibitors of LFA-1
Bioorg.Med.Chem.Lett., 15, 2005
|
|
2W2H
 
 | |
4NST
 
 | | Crystal structure of human Cdk12/Cyclin K in complex with ADP-aluminum fluoride | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, ... | | Authors: | Boesken, C.A, Farnung, L, Anand, K, Geyer, M. | | Deposit date: | 2013-11-29 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure and substrate specificity of human Cdk12/Cyclin K.
Nat Commun, 5, 2014
|
|
5HB2
 
 | |
5CWS
 
 | | Crystal structure of the intact Chaetomium thermophilum Nsp1-Nup49-Nup57 channel nucleoporin heterotrimer bound to its Nic96 nuclear pore complex attachment site | | Descriptor: | Nucleoporin NIC96, Nucleoporin NSP1, Nucleoporin NUP49, ... | | Authors: | Bley, C.J, Petrovic, S, Paduch, M, Lu, V, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2015-07-28 | | Release date: | 2015-10-14 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (3.77 Å) | | Cite: | Architecture of the fungal nuclear pore inner ring complex.
Science, 350, 2015
|
|
5CWT
 
 | |
3OKP
 
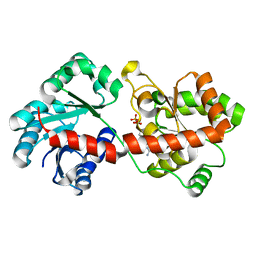 | | Crystal structure of Corynebacterium glutamicum PimB' bound to GDP-Man (orthorhombic crystal form) | | Descriptor: | GDP-mannose-dependent alpha-(1-6)-phosphatidylinositol monomannoside mannosyltransferase, GUANOSINE-5'-DIPHOSPHATE-ALPHA-D-MANNOSE | | Authors: | Batt, S.M, Jabeen, T, Besra, G.S, Futterer, K. | | Deposit date: | 2010-08-25 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Acceptor substrate discrimination in phosphatidyl-myo-inositol mannoside synthesis: structural and mutational analysis of mannosyltransferase Corynebacterium glutamicum PimB'.
J.Biol.Chem., 285, 2010
|
|
3OKA
 
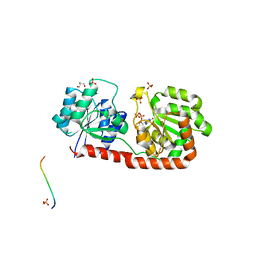 | | Crystal structure of Corynebacterium glutamicum PimB' in complex with GDP-Man (triclinic crystal form) | | Descriptor: | GDP-mannose-dependent alpha-(1-6)-phosphatidylinositol monomannoside mannosyltransferase, GLYCEROL, GUANOSINE-5'-DIPHOSPHATE-ALPHA-D-MANNOSE, ... | | Authors: | Batt, S.M, Jabeen, T, Besra, G.S, Futterer, K. | | Deposit date: | 2010-08-24 | | Release date: | 2010-09-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Acceptor substrate discrimination in phosphatidyl-myo-inositol mannoside synthesis: structural and mutational analysis of mannosyltransferase Corynebacterium glutamicum PimB'.
J.Biol.Chem., 285, 2010
|
|
3OKC
 
 | | Crystal structure of Corynebacterium glutamicum PimB' bound to GDP (orthorhombic crystal form) | | Descriptor: | GDP-mannose-dependent alpha-(1-6)-phosphatidylinositol monomannoside mannosyltransferase, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Batt, S.M, Jabeen, T, Besra, G.S, Futterer, K. | | Deposit date: | 2010-08-24 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Acceptor substrate discrimination in phosphatidyl-myo-inositol mannoside synthesis: structural and mutational analysis of mannosyltransferase Corynebacterium glutamicum PimB'.
J.Biol.Chem., 285, 2010
|
|
4JO9
 
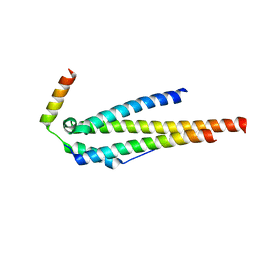 | | Crystal structure of the human Nup49CCS2+3* Nup57CCS3* complex 1:2 stoichiometry | | Descriptor: | Nucleoporin p54, Nucleoporin p58/p45 | | Authors: | Stuwe, T, Bley, C.J, Mayo, D.J, Hoelz, A. | | Deposit date: | 2013-03-18 | | Release date: | 2014-09-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.499 Å) | | Cite: | Architecture of the fungal nuclear pore inner ring complex.
Science, 350, 2015
|
|
4JQ5
 
 | |
4JO7
 
 | | Crystal structure of the human Nup49CCS2+3* Nup57CCS3* complex with 2:2 stoichiometry | | Descriptor: | Nucleoporin p54, Nucleoporin p58/p45 | | Authors: | Stuwe, T, Bley, C.J, Mayo, D.J, Hoelz, A. | | Deposit date: | 2013-03-17 | | Release date: | 2014-09-17 | | Last modified: | 2016-02-03 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | Architecture of the fungal nuclear pore inner ring complex.
Science, 350, 2015
|
|
4JNV
 
 | | Crystal structure of the human Nup57CCS3* coiled-coil segment, space group C2 | | Descriptor: | Nucleoporin p54 | | Authors: | Stuwe, T, Bley, C.J, Mayo, D.J, Hoelz, A. | | Deposit date: | 2013-03-15 | | Release date: | 2014-09-17 | | Last modified: | 2016-02-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Architecture of the fungal nuclear pore inner ring complex.
Science, 350, 2015
|
|
4JNU
 
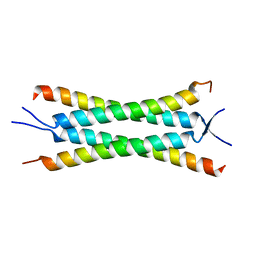 | | Crystal structure of the human Nup57CCS3* coiled-coil segment, space group P21 | | Descriptor: | Nucleoporin p54 | | Authors: | Stuwe, T, Bley, C.J, Mayo, D.J, Hoelz, A. | | Deposit date: | 2013-03-15 | | Release date: | 2014-09-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.445 Å) | | Cite: | Architecture of the fungal nuclear pore inner ring complex.
Science, 350, 2015
|
|
5HAZ
 
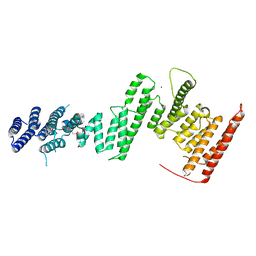 | | Structure of Chaetomium thermophilum Nup170 CTD | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Nucleoporin NUP170, ... | | Authors: | Lin, D.H, Hoelz, A. | | Deposit date: | 2015-12-31 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Architecture of the symmetric core of the nuclear pore.
Science, 352, 2016
|
|
5HB8
 
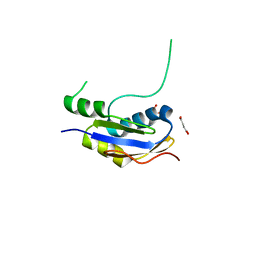 | |
5HB5
 
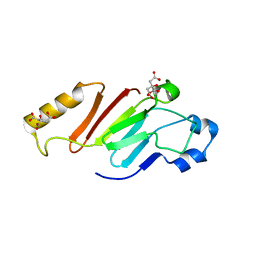 | |
5HB4
 
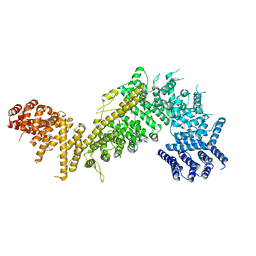 | |
5HAY
 
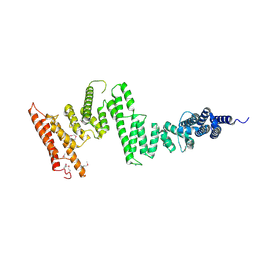 | |
5HAX
 
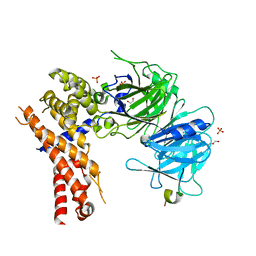 | | Crystal structure of Chaetomium thermophilum Nup170 NTD-Nup53 complex | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Nucleoporin NUP170, ... | | Authors: | Lin, D.H, Mobbs, G, Hoelz, A. | | Deposit date: | 2015-12-31 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Architecture of the symmetric core of the nuclear pore.
Science, 352, 2016
|
|
5HB6
 
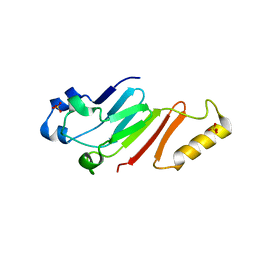 | |
