3J9U
 
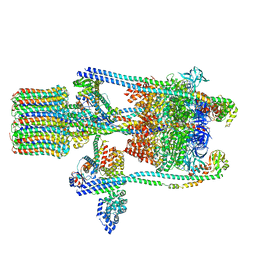 | | Yeast V-ATPase state 2 | | Descriptor: | V-type proton ATPase catalytic subunit A, V-type proton ATPase subunit B, V-type proton ATPase subunit C, ... | | Authors: | Zhao, J, Benlekbir, S, Rubinstein, J.L. | | Deposit date: | 2015-02-23 | | Release date: | 2015-05-13 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Electron cryomicroscopy observation of rotational states in a eukaryotic V-ATPase.
Nature, 521, 2015
|
|
3J9T
 
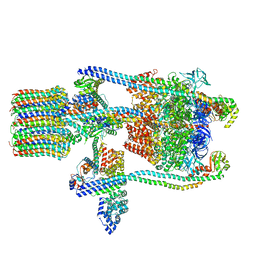 | | Yeast V-ATPase state 1 | | Descriptor: | V-type proton ATPase catalytic subunit A, V-type proton ATPase subunit B, V-type proton ATPase subunit C, ... | | Authors: | Zhao, J, Benlekbir, S, Rubinstein, J.L. | | Deposit date: | 2015-02-23 | | Release date: | 2015-05-13 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Electron cryomicroscopy observation of rotational states in a eukaryotic V-ATPase.
Nature, 521, 2015
|
|
7CFP
 
 | |
7CFQ
 
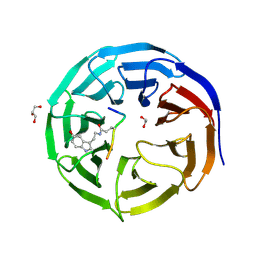 | | Crystal structure of WDR5 in complex with H3K4me3Q5ser peptide | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, H3K4me3Q5ser peptide, ... | | Authors: | Zhao, J, Zhang, X, Zang, J. | | Deposit date: | 2020-06-27 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights into the recognition of histone H3Q5 serotonylation by WDR5.
Sci Adv, 7, 2021
|
|
8CVT
 
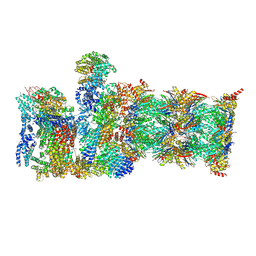 | | Human 19S-20S proteasome, state SD2 | | Descriptor: | 26S protease regulatory subunit 10B, 26S protease regulatory subunit 8, 26S proteasome complex subunit SEM1, ... | | Authors: | Zhao, J. | | Deposit date: | 2022-05-18 | | Release date: | 2022-11-02 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into the human PA28-20S proteasome enabled by efficient tagging and purification of endogenous proteins.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8CVR
 
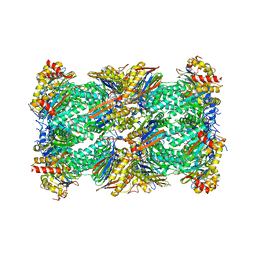 | | Human 20S proteasome with MG-132 | | Descriptor: | N-[(benzyloxy)carbonyl]-L-leucyl-N-[(2S)-4-methyl-1-oxopentan-2-yl]-L-leucinamide, Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, ... | | Authors: | Zhao, J. | | Deposit date: | 2022-05-18 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural insights into the human PA28-20S proteasome enabled by efficient tagging and purification of endogenous proteins.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8CVS
 
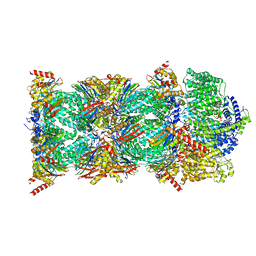 | | Human PA200-20S proteasome with MG-132 | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, N-[(benzyloxy)carbonyl]-L-leucyl-N-[(2S)-4-methyl-1-oxopentan-2-yl]-L-leucinamide, Proteasome activator complex subunit 4, ... | | Authors: | Zhao, J. | | Deposit date: | 2022-05-18 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into the human PA28-20S proteasome enabled by efficient tagging and purification of endogenous proteins.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8CXB
 
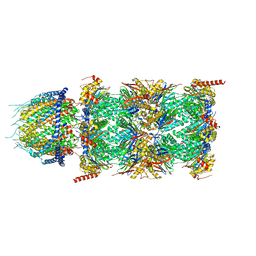 | | Human PA28-20S (PA28-4a3b) | | Descriptor: | Proteasome activator complex subunit 1, Proteasome activator complex subunit 2, Proteasome subunit alpha type-1, ... | | Authors: | Zhao, J, Makhija, S, Huang, B, Cheng, Y. | | Deposit date: | 2022-05-20 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insights into the human PA28-20S proteasome enabled by efficient tagging and purification of endogenous proteins.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8G4G
 
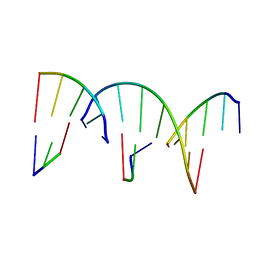 | | Crystal Engineering with One 8-mer DNA | | Descriptor: | DNA (5'-D(*AP*TP*CP*GP*G)-3'), DNA (5'-D(*AP*TP*CP*GP*GP*CP*CP*G)-3'), DNA (5'-D(P*CP*CP*G)-3') | | Authors: | Zhao, J, Zhang, C, Lu, B, Seeman, N.C, Noinaj, N, Sha, R, Mao, C. | | Deposit date: | 2023-02-09 | | Release date: | 2023-05-10 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Divergence and Convergence: Complexity Emerges in Crystal Engineering from an 8-mer DNA.
J.Am.Chem.Soc., 145, 2023
|
|
4JYQ
 
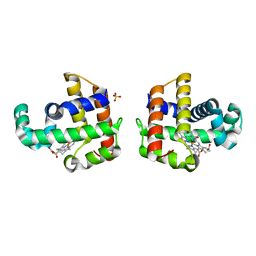 | | DHP-CO crystal structure | | Descriptor: | CARBON MONOXIDE, Dehaloperoxidase A, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Zhao, J, Srajer, V, Franzen, S. | | Deposit date: | 2013-03-31 | | Release date: | 2014-04-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Observation of protein valves for the control of the internal flow of small molecules using time-resolved X-ray crystallography
To be Published
|
|
7FB5
 
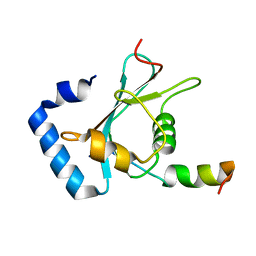 | | Crystal structure of FAM134B/GABARAP complex | | Descriptor: | Gamma-aminobutyric acid receptor-associated protein, Reticulophagy regulator 1 | | Authors: | Zhao, J, Li, J. | | Deposit date: | 2021-07-08 | | Release date: | 2022-03-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | The crystal structure of the FAM134B-GABARAP complex provides mechanistic insights into the selective binding of FAM134 to the GABARAP subfamily.
Febs Open Bio, 12, 2022
|
|
7YAC
 
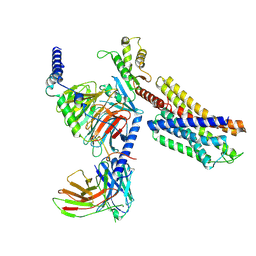 | | Paltusotine-bound SSTR2-Gi complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Zhao, J, Shao, Z. | | Deposit date: | 2022-06-27 | | Release date: | 2023-04-19 | | Last modified: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Prospect of acromegaly therapy: molecular mechanism of clinical drugs octreotide and paltusotine.
Nat Commun, 14, 2023
|
|
7YAE
 
 | | Octreotide-bound SSTR2-Gi complex | | Descriptor: | CHOLESTEROL, DPN-CYS-PHE-DTR-LYS-THR-CYS-THO, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhao, J, Shao, Z. | | Deposit date: | 2022-06-28 | | Release date: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Prospect of acromegaly therapy: molecular mechanism of clinical drugs octreotide and paltusotine.
Nat Commun, 14, 2023
|
|
5KF4
 
 | |
5J2Y
 
 | | Molecular insight into the regulatory mechanism of the quorum-sensing repressor RsaL in Pseudomonas aeruginosa | | Descriptor: | DNA (26-MER), Regulatory protein | | Authors: | Zhao, J, Gan, J, Zhang, J, Kang, H, Kong, W, Zhu, M, Li, F, Song, Y, Qin, J, Liang, H. | | Deposit date: | 2016-03-30 | | Release date: | 2017-04-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of Pseudomonas aeruginosa RsaL bound to promoter DNA reaffirms its role as a global regulator involved in quorum-sensing.
Nucleic Acids Res., 45, 2017
|
|
4ILZ
 
 | | Structural and kinetic study of an internal substrate binding site of dehaloperoxidase-hemoglobin A from Amphitrite ornata | | Descriptor: | 2,4,6-TRIBROMOPHENOL, Dehaloperoxidase A, ISOPROPYL ALCOHOL, ... | | Authors: | de Serrano, V.S, Zhao, J, Zhao, J, Franzen, S. | | Deposit date: | 2013-01-01 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structural and Kinetic Study of an Internal Substrate Binding Site in Dehaloperoxidase-Hemoglobin A from Amphitrite ornata.
Biochemistry, 52, 2013
|
|
6ZT9
 
 | | X-ray structure of mutated arabinofuranosidase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | Authors: | Tandrup, T, Lo Leggio, L, Zhao, J, Bissaro, B, Barbe, S, Andre, I, Dumon, C, O'Donohue, M.J, Faure, R. | | Deposit date: | 2020-07-17 | | Release date: | 2021-02-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Probing the determinants of the transglycosylation/hydrolysis partition in a retaining alpha-l-arabinofuranosidase.
N Biotechnol, 62, 2021
|
|
6ZTA
 
 | | X-ray structure of mutated arabinofuranosidase | | Descriptor: | Alpha-L-arabinofuranosidase | | Authors: | Tandrup, T, Lo Leggio, L, Zhao, J, Bissaro, B, Barbe, S, Andre, I, Dumon, C, O'Donohue, M.J, Faure, R. | | Deposit date: | 2020-07-17 | | Release date: | 2021-02-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Probing the determinants of the transglycosylation/hydrolysis partition in a retaining alpha-l-arabinofuranosidase.
N Biotechnol, 62, 2021
|
|
6ZT7
 
 | | X-ray structure of mutated arabinofuranosidase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | Authors: | Tandrup, T, Lo Leggio, L, Zhao, J, Bissaro, B, Barbe, S, Andre, I, Dumon, C, O'Donohue, M.J, Faure, R. | | Deposit date: | 2020-07-17 | | Release date: | 2021-02-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Probing the determinants of the transglycosylation/hydrolysis partition in a retaining alpha-l-arabinofuranosidase.
N Biotechnol, 62, 2021
|
|
6ZT8
 
 | | X-ray structure of mutated arabinofuranosidase | | Descriptor: | Alpha-L-arabinofuranosidase, CHLORIDE ION, PENTAETHYLENE GLYCOL, ... | | Authors: | Tandrup, T, Lo Leggio, L, Zhao, J, Bissaro, B, Barbe, S, Andre, I, Dumon, C, O'Donohue, M.J, Faure, R. | | Deposit date: | 2020-07-17 | | Release date: | 2021-02-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Probing the determinants of the transglycosylation/hydrolysis partition in a retaining alpha-l-arabinofuranosidase.
N Biotechnol, 62, 2021
|
|
6ZT6
 
 | | X-ray structure of mutated arabinofuranosidase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Alpha-L-arabinofuranosidase | | Authors: | Tandrup, T, Lo Leggio, L, Zhao, J, Bissaro, B, Barbe, S, Andre, I, Dumon, C, O'Donohue, M.J, Faure, R. | | Deposit date: | 2020-07-17 | | Release date: | 2021-02-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Probing the determinants of the transglycosylation/hydrolysis partition in a retaining alpha-l-arabinofuranosidase.
N Biotechnol, 62, 2021
|
|
5H3J
 
 | |
2L54
 
 | | Solution structure of the Zalpha domain mutant of ADAR1 (N43A,Y47A) | | Descriptor: | Double-stranded RNA-specific adenosine deaminase | | Authors: | Zhao, J, Pervushin, K, Feng, S, Droge, P. | | Deposit date: | 2010-10-24 | | Release date: | 2011-01-12 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Alternate rRNA secondary structures as regulators of translation
Nat.Struct.Mol.Biol., 18, 2011
|
|
7VUX
 
 | | Complex structure of PD1 and 609A-Fab | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Huang, H, Zhu, Z, Zhao, J, Jiang, L, Yang, H, Deng, L, Meng, X, Ding, J, Yang, S, Zhao, L, Xu, W, Wang, X. | | Deposit date: | 2021-11-04 | | Release date: | 2021-11-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | A strategy for the efficient construction of anti-PD1-based bispecific antibodies with desired IgG-like properties.
Mabs, 14, 2022
|
|
8ISK
 
 | | Pr conformer of Zea mays phytochrome A1 - ZmphyA1-Pr | | Descriptor: | 3-[5-[[(3~{R},4~{R})-3-ethyl-4-methyl-5-oxidanylidene-3,4-dihydropyrrol-2-yl]methyl]-2-[[5-[(4-ethyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol-2-yl]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, Phytochrome | | Authors: | Zhang, Y, Ma, C, Zhao, J, Gao, N, Wang, J. | | Deposit date: | 2023-03-20 | | Release date: | 2023-08-09 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into plant phytochrome A as a highly sensitized photoreceptor.
Cell Res., 33, 2023
|
|
