5WEU
 
 | | Crystal Structure of H2-Dd with disulfide-linked 10mer peptide | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, Envelope glycoprotein gp160, ... | | Authors: | Jiang, J.S, Natarajan, K, Boyd, L.F, Margulies, D.H. | | Deposit date: | 2017-07-10 | | Release date: | 2017-10-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.584 Å) | | Cite: | Crystal structure of a TAPBPR-MHC I complex reveals the mechanism of peptide editing in antigen presentation.
Science, 358, 2017
|
|
7TUD
 
 | | Crystal structure of HLA-B*44:05 (T73C) with 6mer EEFGRC and dipeptide GL | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, EEFGRC peptide, ... | | Authors: | Jiang, J, Natarajan, K, Kim, E, Boyd, L.F, Margulies, D.H. | | Deposit date: | 2022-02-02 | | Release date: | 2022-09-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural mechanism of tapasin-mediated MHC-I peptide loading in antigen presentation.
Nat Commun, 13, 2022
|
|
7TUG
 
 | | Crystal structure of Tapasin in complex with PaSta2-Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PaSta2 Fab heavy chain, PaSta2 Fab kappa light chain, ... | | Authors: | Jiang, J, Natarajan, K, Taylor, D.K, Boyd, L.F, Margulies, D.H. | | Deposit date: | 2022-02-02 | | Release date: | 2022-09-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Structural mechanism of tapasin-mediated MHC-I peptide loading in antigen presentation.
Nat Commun, 13, 2022
|
|
7TUE
 
 | | Crystal structure of Tapasin in complex with HLA-B*44:05 (T73C) | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B alpha chain, ... | | Authors: | Jiang, J, Natarajan, K, Kim, E, Boyd, L.F, Margulies, D.H. | | Deposit date: | 2022-02-02 | | Release date: | 2022-09-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural mechanism of tapasin-mediated MHC-I peptide loading in antigen presentation.
Nat Commun, 13, 2022
|
|
7TUH
 
 | | Crystal structure of anti-tapasin PaSta2-Fab | | Descriptor: | PaSta2 Fab heavy chain, PaSta2 Fab kappa light chain | | Authors: | Jiang, J, Natarajan, K, Taylor, D.K, Boyd, L.F, Margulies, D.H. | | Deposit date: | 2022-02-02 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural mechanism of tapasin-mediated MHC-I peptide loading in antigen presentation.
Nat Commun, 13, 2022
|
|
7TUC
 
 | | Crystal structure of HLA-B*44:05 (T73C) with 9mer EEFGRAFSF | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Jiang, J, Natarajan, K, Kim, E, Boyd, L.F, Margulies, D.H. | | Deposit date: | 2022-02-02 | | Release date: | 2022-09-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural mechanism of tapasin-mediated MHC-I peptide loading in antigen presentation.
Nat Commun, 13, 2022
|
|
7TUF
 
 | | Crystal structure of Tapasin in complex with PaSta1-Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PaSta1 Fab heavy chain, PaSta1 Fab kappa light chain, ... | | Authors: | Jiang, J, Natarajan, K, Taylor, D.K, Boyd, L.F, Margulies, D.H. | | Deposit date: | 2022-02-02 | | Release date: | 2022-09-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural mechanism of tapasin-mediated MHC-I peptide loading in antigen presentation.
Nat Commun, 13, 2022
|
|
1FM5
 
 | |
7MFV
 
 | | Crystal structure of synthetic nanobody (Sb16) | | Descriptor: | 1,2-ETHANEDIOL, Synthetic Nanobody #16 (Sb16) | | Authors: | Jiang, J, Ahmad, J, Natarajan, K, Boyd, L.F, Margulies, D.H. | | Deposit date: | 2021-04-11 | | Release date: | 2021-06-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of synthetic nanobody-SARS-CoV-2 receptor-binding domain complexes reveal distinct sites of interaction.
J.Biol.Chem., 297, 2021
|
|
7MFU
 
 | | Crystal structure of synthetic nanobody (Sb14+Sb68) complexes with SARS-CoV-2 receptor binding domain | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Spike protein S1, ... | | Authors: | Jiang, J, Ahmad, J, Natarajan, K, Boyd, L.F, Margulies, D.H. | | Deposit date: | 2021-04-11 | | Release date: | 2021-06-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of synthetic nanobody-SARS-CoV-2 receptor-binding domain complexes reveal distinct sites of interaction.
J.Biol.Chem., 297, 2021
|
|
5KD4
 
 | | Crystal Structure of Murine MHC-I H-2Dd in complex with Murine Beta2-Microglobulin and a Variant of Peptide (PVI10) of HIV gp120 MN Isolate (IGPGRAFYVI) | | Descriptor: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, D-D alpha chain, ... | | Authors: | Jiang, J, Natarajan, K, Margulies, D. | | Deposit date: | 2016-06-07 | | Release date: | 2017-10-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Effects of Cross-Presentation, Antigen Processing, and Peptide Binding in HIV Evasion of T Cell Immunity.
J. Immunol., 200, 2018
|
|
1QO3
 
 | |
1JA3
 
 | | Crystal Structure of the Murine NK Cell Inhibitory Receptor Ly-49I | | Descriptor: | MHC class I recognition receptor Ly49I | | Authors: | Dimasi, N, Sawicki, W.M, Reineck, L.A, Li, Y, Natarajan, K, Murgulies, D.H, Mariuzza, A.R. | | Deposit date: | 2001-05-29 | | Release date: | 2002-07-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the Ly49I natural killer cell receptor reveals variability in dimerization mode within the Ly49 family.
J.Mol.Biol., 320, 2002
|
|
5IVX
 
 | | Crystal Structure of B4.2.3 T-Cell Receptor and H2-Dd P18-I10 Complex | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, H-2 class I histocompatibility antigen, ... | | Authors: | Natarajan, K, Jiang, J, Margulies, D. | | Deposit date: | 2016-03-21 | | Release date: | 2017-03-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | An allosteric site in the T-cell receptor C beta domain plays a critical signalling role.
Nat Commun, 8, 2017
|
|
5IW1
 
 | |
5KD7
 
 | | Crystal Structure of Murine MHC-I H-2Dd in complex with Murine Beta2-Microglobulin and a Variant of Peptide (PV9) of HIV gp120 MN Isolate (IGPGRAFYV) | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Jiang, J, Natarajan, K, Margulies, D. | | Deposit date: | 2016-06-07 | | Release date: | 2017-10-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Effects of Cross-Presentation, Antigen Processing, and Peptide Binding in HIV Evasion of T Cell Immunity.
J. Immunol., 200, 2018
|
|
5T7G
 
 | | Crystal Structure of Murine MHC-I H-2Dd in complex with Murine Beta2-Microglobulin and a Variant of Peptide (PT9) of HIV gp120 MN Isolate (IGPGRAFYT) | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, H-2 class I histocompatibility antigen, ... | | Authors: | Jiang, J, Natarajan, K, Margulies, D. | | Deposit date: | 2016-09-04 | | Release date: | 2017-10-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.961 Å) | | Cite: | Effects of Cross-Presentation, Antigen Processing, and Peptide Binding in HIV Evasion of T Cell Immunity.
J. Immunol., 200, 2018
|
|
5TRZ
 
 | | Crystal structure of MHC-I H2-KD complexed with peptides of Mycobacterial tuberculosis (YQSGLSIVM) | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Jiang, J, Natarajan, K, Margulies, D. | | Deposit date: | 2016-10-27 | | Release date: | 2018-05-09 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (2.247 Å) | | Cite: | MHC-restricted Ag85B-specific CD8+T cells are enhanced by recombinant BCG prime and DNA boost immunization in mice.
Eur.J.Immunol., 2019
|
|
5TS1
 
 | | Crystal structure of MHC-I H2-KD complexed with peptides of Mycobacterial tuberculosis (YYQSGLSIV) | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Jiang, J, Natarajan, K, Margulies, D. | | Deposit date: | 2016-10-27 | | Release date: | 2018-05-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | MHC-restricted Ag85B-specific CD8+T cells are enhanced by recombinant BCG prime and DNA boost immunization in mice.
Eur.J.Immunol., 2019
|
|
6B9K
 
 | |
1P4L
 
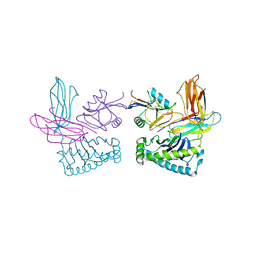 | | Crystal structure of NK receptor Ly49C mutant with its MHC class I ligand H-2Kb | | Descriptor: | Beta-2-microglobulin, LY49-C, MHC CLASS I H-2KB HEAVY CHAIN, ... | | Authors: | Dam, J, Guan, R, Natarajan, K, Dimasi, N, Mariuzza, R.A. | | Deposit date: | 2003-04-23 | | Release date: | 2003-11-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Variable MHC class I engagement by Ly49 natural killer cell receptors demonstrated by the crystal structure of Ly49C bound to H-2K(b).
Nat.Immunol., 4, 2003
|
|
7N0H
 
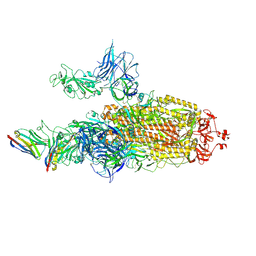 | | CryoEM structure of SARS-CoV-2 spike protein (S-6P, 2-up) in complex with sybodies (Sb45) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Jiang, J, Huang, R, Margulies, D. | | Deposit date: | 2021-05-25 | | Release date: | 2021-06-02 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Structures of synthetic nanobody-SARS-CoV-2 receptor-binding domain complexes reveal distinct sites of interaction.
J.Biol.Chem., 297, 2021
|
|
7N0G
 
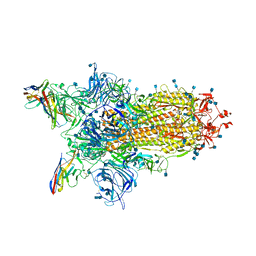 | | CryoEm structure of SARS-CoV-2 spike protein (S-6P, 1-up) in complex with sybodies (Sb45) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Jiang, J, Huang, R, Margulies, D. | | Deposit date: | 2021-05-25 | | Release date: | 2021-06-02 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Structures of synthetic nanobody-SARS-CoV-2 receptor-binding domain complexes reveal distinct sites of interaction.
J.Biol.Chem., 297, 2021
|
|
2H1P
 
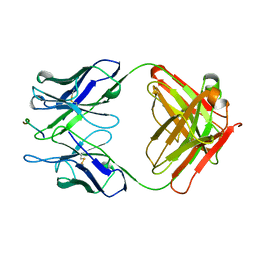 | | THE THREE-DIMENSIONAL STRUCTURES OF A POLYSACCHARIDE BINDING ANTIBODY TO CRYPTOCOCCUS NEOFORMANS AND ITS COMPLEX WITH A PEPTIDE FROM A PHAGE DISPLAY LIBRARY: IMPLICATIONS FOR THE IDENTIFICATION OF PEPTIDE MIMOTOPES | | Descriptor: | 2H1, PA1 | | Authors: | Young, A.C.M, Valadon, P, Casadevall, A, Scharff, M.D, Sacchettini, J.C. | | Deposit date: | 1997-11-12 | | Release date: | 1998-01-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The three-dimensional structures of a polysaccharide binding antibody to Cryptococcus neoformans and its complex with a peptide from a phage display library: implications for the identification of peptide mimotopes.
J.Mol.Biol., 274, 1997
|
|
7U9O
 
 | | SARS-CoV-2 spike trimer RBD in complex with Fab NE12 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, NE12 Fab heavy chain, NE12 Fab light chain, ... | | Authors: | Tsybovsky, Y, Kwong, P.D, Farci, P. | | Deposit date: | 2022-03-11 | | Release date: | 2022-11-23 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Potent monoclonal antibodies neutralize Omicron sublineages and other SARS-CoV-2 variants.
Cell Rep, 41, 2022
|
|
