3TJD
 
 | | co-crystal structure of Jak2 with thienopyridine 19 | | Descriptor: | 4-amino-2-[4-(tert-butylsulfamoyl)phenyl]-N-methylthieno[3,2-c]pyridine-7-carboxamide, Tyrosine-protein kinase JAK2 | | Authors: | Huang, X. | | Deposit date: | 2011-08-24 | | Release date: | 2011-11-30 | | Last modified: | 2011-12-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery of potent and highly selective thienopyridine janus kinase 2 inhibitors.
J.Med.Chem., 54, 2011
|
|
6LKC
 
 | | Crystal structure of PfaD from Shewanella piezotolerans in complex with FMN | | Descriptor: | CALCIUM ION, FLAVIN MONONUCLEOTIDE, GLYCEROL, ... | | Authors: | Zhang, M.L, Li, Q, Meng, S.S, Guo, L.J, He, L, Huang, J.Z, Li, L, Zhang, H.D. | | Deposit date: | 2019-12-19 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Structural Insights into the Trans -Acting Enoyl Reductase in the Biosynthesis of Long-Chain Polyunsaturated Fatty Acids in Shewanella piezotolerans .
J.Agric.Food Chem., 69, 2021
|
|
5ISY
 
 | | Crystal structure of Nudix family protein with NAD | | Descriptor: | NADH pyrophosphatase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ZINC ION | | Authors: | Zhang, D, Guan, Z, Zou, T, Yin, P. | | Deposit date: | 2016-03-15 | | Release date: | 2016-09-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.354 Å) | | Cite: | Structural basis of prokaryotic NAD-RNA decapping by NudC
Cell Res., 26, 2016
|
|
7YPX
 
 | | Cyanophage Pam3 fiber | | Descriptor: | Pam3 tail fiber proreins, tail fiber chaperone | | Authors: | Wei, Z.L, Jiang, Y.L, Zhou, C.Z. | | Deposit date: | 2022-08-04 | | Release date: | 2022-11-09 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Structural Insights into the Chaperone-Assisted Assembly of a Simplified Tail Fiber of the Myocyanophage Pam3.
Viruses, 14, 2022
|
|
4ZZH
 
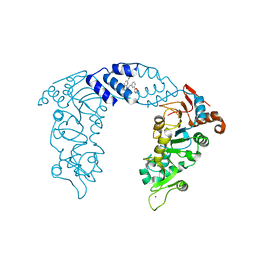 | | SIRT1/Activator Complex | | Descriptor: | (4S)-N-[3-(1,3-oxazol-5-yl)phenyl]-7-[3-(trifluoromethyl)phenyl]-3,4-dihydro-1,4-methanopyrido[2,3-b][1,4]diazepine-5(2H)-carboxamide, NAD-dependent protein deacetylase sirtuin-1, ZINC ION | | Authors: | Dai, H. | | Deposit date: | 2015-05-22 | | Release date: | 2015-07-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.1001 Å) | | Cite: | Crystallographic structure of a small molecule SIRT1 activator-enzyme complex.
Nat Commun, 6, 2015
|
|
4KMH
 
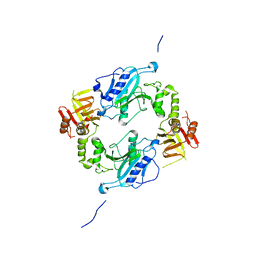 | |
6BBV
 
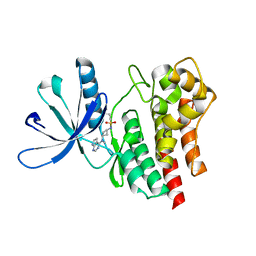 | | Crystal Structure of JAK2 in complex with compound 25 | | Descriptor: | N-{cis-3-[methyl(7H-pyrrolo[2,3-d]pyrimidin-4-yl)amino]cyclobutyl}propane-1-sulfonamide, Tyrosine-protein kinase JAK2 | | Authors: | Han, S. | | Deposit date: | 2017-10-19 | | Release date: | 2018-01-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification of N-{cis-3-[Methyl(7H-pyrrolo[2,3-d]pyrimidin-4-yl)amino]cyclobutyl}propane-1-sulfonamide (PF-04965842): A Selective JAK1 Clinical Candidate for the Treatment of Autoimmune Diseases.
J. Med. Chem., 61, 2018
|
|
6C4Y
 
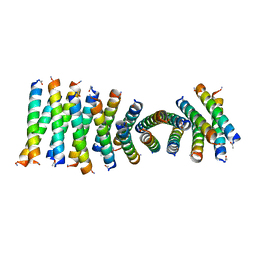 | | Cross-alpha Amyloid-like Structure alphaAmG | | Descriptor: | Cross-alpha Amyloid-like Structure alphaAmG | | Authors: | Liu, L, Zhang, S.Q. | | Deposit date: | 2018-01-13 | | Release date: | 2018-08-15 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Designed peptides that assemble into cross-alpha amyloid-like structures.
Nat. Chem. Biol., 14, 2018
|
|
4KM9
 
 | |
4M6B
 
 | | Crystal structure of yeast Swr1-Z domain in complex with H2A.Z-H2B dimer | | Descriptor: | Chimera protein of Histone H2B.1 and Histone H2A.Z, Helicase SWR1 | | Authors: | Hong, J.J, Feng, H.Q, Wang, F, Ranjan, A, Chen, J.H, Jiang, J.S, Girlando, R, Xiao, T.S, Wu, C, Bai, Y.W. | | Deposit date: | 2013-08-09 | | Release date: | 2014-02-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | The Catalytic Subunit of the SWR1 Remodeler Is a Histone Chaperone for the H2A.Z-H2B Dimer.
Mol.Cell, 53, 2014
|
|
2Z8N
 
 | | Structural basis for the catalytic mechanism of phosphothreonine lyase | | Descriptor: | 27.5 kDa virulence protein, SULFATE ION | | Authors: | Chen, L, Wang, H, Gu, L, Huang, N, Zhou, J.M, Chai, J. | | Deposit date: | 2007-09-07 | | Release date: | 2007-12-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the catalytic mechanism of phosphothreonine lyase.
Nat.Struct.Mol.Biol., 15, 2008
|
|
2Z8M
 
 | | Structural basis for the catalytic mechanism of phosphothreonine lyase | | Descriptor: | 27.5 kDa virulence protein | | Authors: | Chen, L, Wang, H, Gu, L, Huang, N, Zhou, J.M, Chai, J. | | Deposit date: | 2007-09-07 | | Release date: | 2007-12-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the catalytic mechanism of phosphothreonine lyase.
Nat.Struct.Mol.Biol., 15, 2008
|
|
8HFQ
 
 | | Cryo-EM structure of CpcL-PBS from cyanobacterium Synechocystis sp. PCC 6803 | | Descriptor: | C-phycocyanin alpha subunit, C-phycocyanin beta subunit, Ferredoxin--NADP reductase, ... | | Authors: | Zheng, L, Zhang, Z, Wang, H, Zheng, Z, Gao, N, Zhao, J. | | Deposit date: | 2022-11-11 | | Release date: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Cryo-EM and femtosecond spectroscopic studies provide mechanistic insight into the energy transfer in CpcL-phycobilisomes.
Nat Commun, 14, 2023
|
|
4XAQ
 
 | | mGluR2 ECD and mGluR3 ECD with ligands | | Descriptor: | (1S,2S,5R,6S)-2-aminobicyclo[3.1.0]hexane-2,6-dicarboxylic acid, CHLORIDE ION, Metabotropic glutamate receptor 2, ... | | Authors: | Clawson, D.K. | | Deposit date: | 2014-12-15 | | Release date: | 2015-02-04 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Synthesis and Pharmacological Characterization of C4-Disubstituted Analogs of 1S,2S,5R,6S-2-Aminobicyclo[3.1.0]hexane-2,6-dicarboxylate: Identification of a Potent, Selective Metabotropic Glutamate Receptor Agonist and Determination of Agonist-Bound Human mGlu2 and mGlu3 Amino Terminal Domain Structures.
J.Med.Chem., 58, 2015
|
|
8C0T
 
 | | NRS 1.2: Fluorescent Sensors for Imaging Interstitial Calcium | | Descriptor: | CALCIUM ION, SULFATE ION, mNeonGreen,Optimized Ratiometric Calcium Sensor | | Authors: | Basquin, J, Griesbeck, O, Valiente-Gabioud, A. | | Deposit date: | 2022-12-19 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Fluorescent sensors for imaging of interstitial calcium.
Nat Commun, 14, 2023
|
|
4XAR
 
 | | mGluR2 ECD and mGluR3 ECD complex with ligands | | Descriptor: | (1S,2S,5R,6S)-2-aminobicyclo[3.1.0]hexane-2,6-dicarboxylic acid, IODIDE ION, Metabotropic glutamate receptor 3 | | Authors: | Clawson, D.K. | | Deposit date: | 2014-12-15 | | Release date: | 2015-02-11 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Synthesis and Pharmacological Characterization of C4-Disubstituted Analogs of 1S,2S,5R,6S-2-Aminobicyclo[3.1.0]hexane-2,6-dicarboxylate: Identification of a Potent, Selective Metabotropic Glutamate Receptor Agonist and Determination of Agonist-Bound Human mGlu2 and mGlu3 Amino Terminal Domain Structures.
J.Med.Chem., 58, 2015
|
|
6ISO
 
 | | Human SIRT3 Recognizing H3K4cr | | Descriptor: | (2E)-BUT-2-ENAL, ARG-THR-LYS-GLN-THR-ALA-ARG, GLYCEROL, ... | | Authors: | Wang, Y, Hao, Q. | | Deposit date: | 2018-11-17 | | Release date: | 2019-01-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Identification of 'erasers' for lysine crotonylated histone marks using a chemical proteomics approach.
Elife, 3, 2014
|
|
6YFZ
 
 | |
7VRD
 
 | |
5GPK
 
 | | Crystal structure of Ccp1 mutant | | Descriptor: | Putative nucleosome assembly protein C36B7.08c | | Authors: | Yin, F, Gao, F, Chen, Y. | | Deposit date: | 2016-08-03 | | Release date: | 2016-11-30 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | Ccp1 Homodimer Mediates Chromatin Integrity by Antagonizing CENP-A Loading
Mol.Cell, 64, 2016
|
|
7V67
 
 | |
6FTR
 
 | | Serial Femtosecond Crystallography at Megahertz pulse rates | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Wiedorn, M.O, Oberthuer, D, Barty, A, Chapman, H.N. | | Deposit date: | 2018-02-23 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.76000106 Å) | | Cite: | Megahertz serial crystallography.
Nat Commun, 9, 2018
|
|
7KPT
 
 | | Crystal structure of CtdE in complex with FAD and substrate 4 | | Descriptor: | (6aR,7aS,11S,13aS)-6,6,11-trimethyl-4-(3-methylbut-2-en-1-yl)-6,6a,7,8,9,10,11,14-octahydro-5H,13H-13a,7a-(epiminomethano)quinolizino[2,3-b]carbazol-16-one, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Zhao, B, Hu, L. | | Deposit date: | 2020-11-12 | | Release date: | 2021-06-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structural basis of the stereoselective formation of the spirooxindole ring in the biosynthesis of citrinadins.
Nat Commun, 12, 2021
|
|
7KPQ
 
 | | Crystal structure of CtdE in complex with FAD | | Descriptor: | FAD-dependent monooxygenase CtdE, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Zhao, B, Hu, L. | | Deposit date: | 2020-11-12 | | Release date: | 2021-06-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of the stereoselective formation of the spirooxindole ring in the biosynthesis of citrinadins.
Nat Commun, 12, 2021
|
|
6YG4
 
 | |
