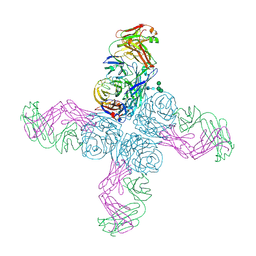
6N6B
 
 | |
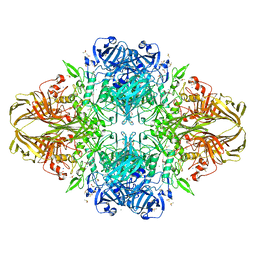
1JYW
 
 | |
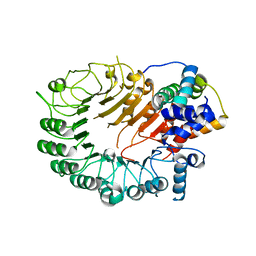
5HYW
 
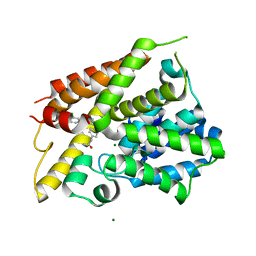
 | | The crystal structure of the D3-ASK1 complex | | Descriptor: | F-box/LRR-repeat MAX2 homolog, SKP1-like protein 1A | | Authors: | Yao, R.F, Ming, Z.H, Yan, L.M, Rao, Z.H, Lou, Z.Y, Xie, D.X. | | Deposit date: | 2016-02-02 | | Release date: | 2016-08-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | DWARF14 is a non-canonical hormone receptor for strigolactone
Nature, 536, 2016
|
|
6NJH
 
 | | Crystal Structure of the PDE4D Catalytic Domain and UCR2 Regulatory Helix with T-48 | | Descriptor: | 2-(4-{[4-(3-chlorophenyl)-6-ethyl-1,3,5-triazin-2-yl]amino}phenyl)acetamide, MAGNESIUM ION, ZINC ION, ... | | Authors: | Fox III, D, Fairman, J.W, Gurney, M.E. | | Deposit date: | 2019-01-03 | | Release date: | 2019-05-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Design and Synthesis of Selective Phosphodiesterase 4D (PDE4D) Allosteric Inhibitors for the Treatment of Fragile X Syndrome and Other Brain Disorders.
J.Med.Chem., 62, 2019
|
|
6NFT
 
 | | Structure of USP5 zinc-finger ubiquitin binding domain co-crystallized with (4-oxoquinazolin-3(4H)-yl)acetic acid | | Descriptor: | (4-oxoquinazolin-3(4H)-yl)acetic acid, 1,2-ETHANEDIOL, UNKNOWN ATOM OR ION, ... | | Authors: | Harding, R.J, Mann, M.K, Tempel, W, Bountra, C, Arrowmsmith, C.M, Edwards, A.M, Schapira, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-12-20 | | Release date: | 2019-01-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Discovery of Small Molecule Antagonists of the USP5 Zinc Finger Ubiquitin-Binding Domain.
J.Med.Chem., 62, 2019
|
|
6NJJ
 
 | | Crystal Structure of the PDE4D Catalytic Domain and UCR2 Regulatory Helix with BPN14770 | | Descriptor: | (4-{[2-(3-chlorophenyl)-6-(trifluoromethyl)pyridin-4-yl]methyl}phenyl)acetic acid, 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Fox III, D, Fairman, J.W, Gurney, M.E. | | Deposit date: | 2019-01-03 | | Release date: | 2019-05-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design and Synthesis of Selective Phosphodiesterase 4D (PDE4D) Allosteric Inhibitors for the Treatment of Fragile X Syndrome and Other Brain Disorders.
J.Med.Chem., 62, 2019
|
|
6NJI
 
 | | Crystal Structure of the PDE4D Catalytic Domain and UCR2 Regulatory Helix with T-49 | | Descriptor: | 2-(4-{[4-(3-chlorophenyl)-6-ethyl-1,3,5-triazin-2-yl]amino}phenyl)ethan-1-ol, MAGNESIUM ION, ZINC ION, ... | | Authors: | Fox III, D, Fairman, J.W, Gurney, M.E. | | Deposit date: | 2019-01-03 | | Release date: | 2019-05-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Design and Synthesis of Selective Phosphodiesterase 4D (PDE4D) Allosteric Inhibitors for the Treatment of Fragile X Syndrome and Other Brain Disorders.
J.Med.Chem., 62, 2019
|
|
6NY4
 
 | |
6OR7
 
 | | Structure of HIV-1 Reverse Transcriptase (RT) in complex with DNA AND (-)FTC-TP | | Descriptor: | DNA Primer 20-mer, DNA template 27-mer, MAGNESIUM ION, ... | | Authors: | Bertoletti, N, Chan, A.H, Anderson, K.S. | | Deposit date: | 2019-04-29 | | Release date: | 2019-07-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structural insights into the recognition of nucleoside reverse transcriptase inhibitors by HIV-1 reverse transcriptase: First crystal structures with reverse transcriptase and the active triphosphate forms of lamivudine and emtricitabine.
Protein Sci., 28, 2019
|
|
6OUN
 
 | | Structure of HIV-1 Reverse Transcriptase (RT) in complex with dsDNA and (-)3TC-TP | | Descriptor: | DNA primer 20-mer, DNA template 27-mer, Lamivudine Triphosphate, ... | | Authors: | Bertoletti, N, Anderson, K.S. | | Deposit date: | 2019-05-04 | | Release date: | 2019-07-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.656 Å) | | Cite: | Structural insights into the recognition of nucleoside reverse transcriptase inhibitors by HIV-1 reverse transcriptase: First crystal structures with reverse transcriptase and the active triphosphate forms of lamivudine and emtricitabine.
Protein Sci., 28, 2019
|
|
6OTZ
 
 | | Structure of HIV-1 Reverse Transcriptase (RT) in complex with dsDNA and (+)FTC-TP | | Descriptor: | DNA Primer 20-mer, DNA template 27-mer, GLYCEROL, ... | | Authors: | Bertoletti, N, Anderson, K.S. | | Deposit date: | 2019-05-03 | | Release date: | 2019-07-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.857 Å) | | Cite: | Structural insights into the recognition of nucleoside reverse transcriptase inhibitors by HIV-1 reverse transcriptase: First crystal structures with reverse transcriptase and the active triphosphate forms of lamivudine and emtricitabine.
Protein Sci., 28, 2019
|
|
6OVZ
 
 | | Crystal structure of the New Delhi metallo-beta-lactamase-1 adduct with a lysine-targeted affinity label | | Descriptor: | Beta-lactamase, CALCIUM ION, ZINC ION, ... | | Authors: | Monzingo, A.F, Fast, W, Thomas, P.W. | | Deposit date: | 2019-05-08 | | Release date: | 2019-06-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.017 Å) | | Cite: | A Lysine-Targeted Affinity Label for Serine-beta-Lactamase Also Covalently Modifies New Delhi Metallo-beta-lactamase-1 (NDM-1).
Biochemistry, 58, 2019
|
|
6P2G
 
 | | Structure of HIV-1 Reverse Transcriptase (RT) in complex with dsDNA and D-ddCTP | | Descriptor: | 2',3'-DIDEOXYCYTIDINE 5'-TRIPHOSPHATE, DNA Primer 20-mer, DNA template 27-mer, ... | | Authors: | Bertoletti, N, Anderson, K.S. | | Deposit date: | 2019-05-21 | | Release date: | 2019-07-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Structural insights into the recognition of nucleoside reverse transcriptase inhibitors by HIV-1 reverse transcriptase: First crystal structures with reverse transcriptase and the active triphosphate forms of lamivudine and emtricitabine.
Protein Sci., 28, 2019
|
|
5HZG
 
 | | The crystal structure of the strigolactone-induced AtD14-D3-ASK1 complex | | Descriptor: | (2Z)-2-methylbut-2-ene-1,4-diol, F-box/LRR-repeat MAX2 homolog, SKP1-like protein 1A, ... | | Authors: | Yao, R.F, Ming, Z.H, Yan, L.M, Rao, Z.H, Lou, Z.Y, Xie, D.X. | | Deposit date: | 2016-02-02 | | Release date: | 2016-08-03 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | DWARF14 is a non-canonical hormone receptor for strigolactone
Nature, 536, 2016
|
|
4DH6
 
 | | Structure of Bace-1 (Beta-Secretase) in Complex with (2R)-N-((2S,3R)-1-(benzo[d][1,3]dioxol-5-yl)-3-hydroxy-4-((S)-6'-neopentyl-3',4'-dihydrospiro[cyclobutane-1,2'-pyrano[2,3-b]pyridine]-4'-ylamino)butan-2-yl)-2-methoxypropanamide | | Descriptor: | (2R)-N-[(2S,3R)-1-(1,3-benzodioxol-5-yl)-4-{[(4'S)-6'-(2,2-dimethylpropyl)-3',4'-dihydrospiro[cyclobutane-1,2'-pyrano[2,3-b]pyridin]-4'-yl]amino}-3-hydroxybutan-2-yl]-2-methoxypropanamide, Beta-secretase 1, GLYCEROL, ... | | Authors: | Sickmier, E.A. | | Deposit date: | 2012-01-27 | | Release date: | 2012-04-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Design and preparation of a potent series of hydroxyethylamine containing beta-secretase inhibitors that demonstrate robust reduction of central beta-amyloid.
J.Med.Chem., 55, 2012
|
|
4F7B
 
 | | Structure of the lysosomal domain of limp-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Neculai, D, Ravichandran, M, Seitova, A, Neculai, M, Pizzaro, J.C, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Dhe-Paganon, D, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-05-15 | | Release date: | 2013-10-09 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of LIMP-2 provides functional insights with implications for SR-BI and CD36.
Nature, 504, 2013
|
|
4DI2
 
 | | Crystal structure of BACE1 in complex with hydroxyethylamine inhibitor 37 | | Descriptor: | (2R)-N-{(2S,3R)-4-{[(4'S)-6'-(2,2-dimethylpropyl)-3',4'-dihydrospiro[cyclobutane-1,2'-pyrano[2,3-b]pyridin]-4'-yl]amino}-3-hydroxy-1-[3-(1,3-thiazol-2-yl)phenyl]butan-2-yl}-2-methoxypropanamide, Beta-secretase 1, GLYCEROL | | Authors: | Whittington, D.A, Long, A.M. | | Deposit date: | 2012-01-30 | | Release date: | 2012-10-10 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design and synthesis of potent, orally efficacious hydroxyethylamine derived beta-site amyloid precursor protein cleaving enzyme (BACE1) inhibitors.
J.Med.Chem., 55, 2012
|
|
4EK1
 
 | | Crystal Structure of Electron-Spin Labeled Cytochrome P450cam | | Descriptor: | CAMPHOR, Camphor 5-monooxygenase, POTASSIUM ION, ... | | Authors: | Lee, Y.-T, Goodin, D.B. | | Deposit date: | 2012-04-08 | | Release date: | 2012-07-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Double electron-electron resonance shows cytochrome P450cam undergoes a conformational change in solution upon binding substrate.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4EYJ
 
 | | MAPK13 Complex with inhibitor | | Descriptor: | 1-(3-tert-butyl-1-methyl-1H-pyrazol-5-yl)-3-[4-(pyridin-4-yloxy)phenyl]urea, Mitogen-activated protein kinase 13 | | Authors: | Miller, C.A, Brett, T.J. | | Deposit date: | 2012-05-01 | | Release date: | 2012-12-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | IL-13-induced airway mucus production is attenuated by MAPK13 inhibition.
J.Clin.Invest., 122, 2012
|
|
4GBD
 
 | | Crystal Structure Of Adenosine Deaminase From Pseudomonas Aeruginosa Pao1 with bound Zn and methylthio-coformycin | | Descriptor: | (8R)-3-(5-S-methyl-5-thio-beta-D-ribofuranosyl)-3,6,7,8-tetrahydroimidazo[4,5-d][1,3]diazepin-8-ol, PHOSPHATE ION, Putative uncharacterized protein, ... | | Authors: | Ho, M, Guan, R, Almo, S.C, Schramm, V.L. | | Deposit date: | 2012-07-27 | | Release date: | 2013-06-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.975 Å) | | Cite: | Methylthioadenosine deaminase in an alternative quorum sensing pathway in Pseudomonas aeruginosa.
Biochemistry, 51, 2012
|
|
4EYM
 
 | | MAPK13 complex with inhibitor | | Descriptor: | 2-(morpholin-4-yl)-N-[4-(pyridin-4-yloxy)phenyl]pyridine-4-carboxamide, Mitogen-activated protein kinase 13 | | Authors: | Miller, C.A, Brett, T.J. | | Deposit date: | 2012-05-01 | | Release date: | 2012-12-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.353 Å) | | Cite: | IL-13-induced airway mucus production is attenuated by MAPK13 inhibition.
J.Clin.Invest., 122, 2012
|
|
1TES
 
 | | OXYGEN BINDING MUSCLE PROTEIN | | Descriptor: | METHYLETHYLAMINE, MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Smith, R.D, Phillips Jr, G.N, Olson, J.S. | | Deposit date: | 1996-05-06 | | Release date: | 1996-11-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism of NO-induced oxidation of myoglobin and hemoglobin.
Biochemistry, 35, 1996
|
|
4GS6
 
 | | Irreversible Inhibition of TAK1 Kinase by 5Z-7-Oxozeaenol | | Descriptor: | (3S,5Z,8S,9S,11E)-8,9,16-trihydroxy-14-methoxy-3-methyl-3,4,9,10-tetrahydro-1H-2-benzoxacyclotetradecine-1,7(8H)-dione, 1,2-ETHANEDIOL, Tak1-Tab1 fusion protein | | Authors: | Larsen, N.A, Ferguson, A.D, Wu, J. | | Deposit date: | 2012-08-27 | | Release date: | 2013-01-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanism and In Vitro Pharmacology of TAK1 Inhibition by (5Z)-7-Oxozeaenol.
Acs Chem.Biol., 8, 2013
|
|
6XFT
 
 | |
6YD1
 
 | | SaFtsZ-DFMBA | | Descriptor: | 2,6-difluoro-3-methoxybenzamide, Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Fernandez-Tornero, C, Ruiz, F.M, Andreu, J.M. | | Deposit date: | 2020-03-20 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Targeting the FtsZ Allosteric Binding Site with a Novel Fluorescence Polarization Screen, Cytological and Structural Approaches for Antibacterial Discovery.
J.Med.Chem., 64, 2021
|
|
