4WS2
 
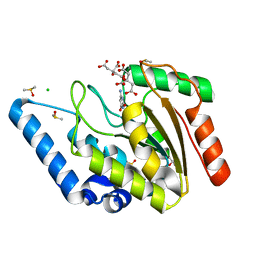 | | Crystal structure of Mycobacterium tuberculosis uracil-DNA glycosylase in complex with 6-aminouracil, Form I | | Descriptor: | 6-aminopyrimidine-2,4(3H,5H)-dione, CHLORIDE ION, CITRIC ACID, ... | | Authors: | Arif, S.M, Geethanandan, K, Mishra, P, Surolia, A, Varshney, U, Vijayan, M. | | Deposit date: | 2014-10-25 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Structural plasticity in Mycobacterium tuberculosis uracil-DNA glycosylase (MtUng) and its functional implications.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4WS7
 
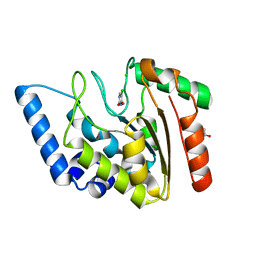 | | Crystal structure of Mycobacterium tuberculosis uracil-DNA glycosylase in complex with 5-chlorouracil, Form II | | Descriptor: | 1,2-ETHANEDIOL, 5-chloropyrimidine-2,4(1H,3H)-dione, CHLORIDE ION, ... | | Authors: | Arif, S.M, Geethanandan, K, Mishra, P, Surolia, A, Varshney, U, Vijayan, M. | | Deposit date: | 2014-10-25 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural plasticity in Mycobacterium tuberculosis uracil-DNA glycosylase (MtUng) and its functional implications.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4WSB
 
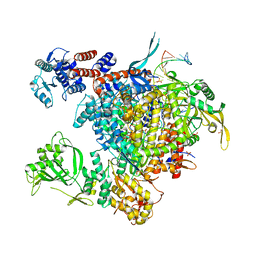 | | Bat Influenza A polymerase with bound vRNA promoter | | Descriptor: | Influenza A polymerase vRNA promoter 3' end, Influenza A polymerase vRNA promoter 5' end, PHOSPHATE ION, ... | | Authors: | Cusack, S, Pflug, A, Guilligay, D, Reich, S. | | Deposit date: | 2014-10-26 | | Release date: | 2014-11-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural insight into cap-snatching and RNA synthesis by influenza polymerase.
Nature, 516, 2014
|
|
4WVB
 
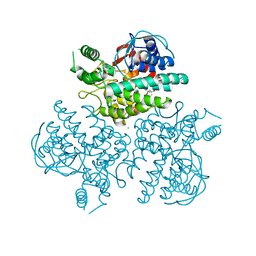 | | Crystal structure of GH63 mannosylglycerate hydrolase from Thermus thermophilus HB8 in complex with glucose | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Uncharacterized protein, ... | | Authors: | Miyazaki, T, Ichikawa, M, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2014-11-05 | | Release date: | 2015-03-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure and substrate-binding mode of GH63 mannosylglycerate hydrolase from Thermus thermophilus HB8.
J.Struct.Biol., 190, 2015
|
|
4YPS
 
 | | (R)-2-Phenylpyrrolidine Substitute Imidazopyridazines: a New Class of Potent and Selective Pan-TRK Inhibitors | | Descriptor: | 4-{6-[(3R)-3-(3-fluorophenyl)morpholin-4-yl]imidazo[1,2-b]pyridazin-3-yl}benzonitrile, High affinity nerve growth factor receptor, SULFATE ION | | Authors: | Kreusch, A, Rucker, P, Molteni, V, Loren, J. | | Deposit date: | 2015-03-13 | | Release date: | 2015-06-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1012 Å) | | Cite: | (R)-2-Phenylpyrrolidine Substituted Imidazopyridazines: A New Class of Potent and Selective Pan-TRK Inhibitors.
Acs Med.Chem.Lett., 6, 2015
|
|
4WT3
 
 | | The N-terminal domain of Rubisco Accumulation Factor 1 from Arabidopsis thaliana | | Descriptor: | Rubisco Accumulation Factor 1, isoform 2 | | Authors: | Hauser, T, Bhat, J.Y, Milicic, G, Wendler, P, Hartl, F.U, Bracher, A, Hayer-Hartl, M. | | Deposit date: | 2014-10-29 | | Release date: | 2015-07-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.954 Å) | | Cite: | Structure and mechanism of the Rubisco-assembly chaperone Raf1.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4WT4
 
 | | The C-terminal domain of Rubisco Accumulation Factor 1 from Arabidopsis thaliana, crystal form I | | Descriptor: | PHOSPHATE ION, Rubisco Accumulation Factor 1, isoform 2 | | Authors: | Hauser, T, Bhat, J.Y, Milicic, G, Wendler, P, Hartl, F.U, Bracher, A, Hayer-Hartl, M. | | Deposit date: | 2014-10-29 | | Release date: | 2015-07-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Structure and mechanism of the Rubisco-assembly chaperone Raf1.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4WT5
 
 | | The C-terminal domain of Rubisco Accumulation Factor 1 from Arabidopsis thaliana, crystal form II | | Descriptor: | Rubisco Accumulation Factor 1, isoform 2 | | Authors: | Hauser, T, Bhat, J.Y, Milicic, G, Wendler, P, Hartl, F.U, Bracher, A, Hayer-Hartl, M. | | Deposit date: | 2014-10-29 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.568 Å) | | Cite: | Structure and mechanism of the Rubisco-assembly chaperone Raf1.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4WT7
 
 | | Crystal structure of an ABC transporter solute binding protein (IPR025997) from Agrobacterium vitis (Avi_5165, Target EFI-511223) with bound allitol | | Descriptor: | ABC transporter substrate binding protein (Ribose), CHLORIDE ION, D-allitol | | Authors: | Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-10-29 | | Release date: | 2014-11-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of an ABC transporter solute binding protein (IPR025997) from Agrobacterium vitis (Avi_5165, Target EFI-511223) with bound allitol
To be published
|
|
4WT8
 
 | | Crystal Structure of bactobolin A bound to 70S ribosome-tRNA complex | | Descriptor: | 23S rRNA (2899-MER), 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Amunts, A, Fiedorczuk, K, Ramakrishnan, V. | | Deposit date: | 2014-10-29 | | Release date: | 2015-01-21 | | Last modified: | 2018-07-18 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Bactobolin A Binds to a Site on the 70S Ribosome Distinct from Previously Seen Antibiotics.
J.Mol.Biol., 427, 2015
|
|
4WJ4
 
 | | Crystal structure of non-discriminating aspartyl-tRNA synthetase from Pseudomonas aeruginosa complexed with tRNA(Asn) and aspartic acid | | Descriptor: | 76mer-tRNA, ASPARTIC ACID, Aspartate--tRNA(Asp/Asn) ligase | | Authors: | Suzuki, T, Nakamura, A, Kato, K, Tanaka, I, Yao, M. | | Deposit date: | 2014-09-29 | | Release date: | 2014-12-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.294 Å) | | Cite: | Structure of the Pseudomonas aeruginosa transamidosome reveals unique aspects of bacterial tRNA-dependent asparagine biosynthesis
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5KPM
 
 | | Glycogen Synthase Kinase 3 beta Complexed with BRD3731 | | Descriptor: | (4~{S})-3-(2,2-dimethylpropyl)-4,7,7-trimethyl-4-phenyl-2,6,8,9-tetrahydropyrazolo[3,4-b]quinolin-5-one, Glycogen synthase kinase-3 beta | | Authors: | Lakshminarasimhan, D, White, A, Nadupalli, A, Suto, R.K. | | Deposit date: | 2016-07-04 | | Release date: | 2018-03-14 | | Last modified: | 2018-03-21 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Exploiting an Asp-Glu "switch" in glycogen synthase kinase 3 to design paralog-selective inhibitors for use in acute myeloid leukemia.
Sci Transl Med, 10, 2018
|
|
4WKY
 
 | | Streptomcyes albus JA3453 oxazolomycin ketosynthase domain OzmN KS2 | | Descriptor: | 1,2-ETHANEDIOL, Beta-ketoacyl synthase, GLYCEROL, ... | | Authors: | Cuff, M.E, Mack, J.C, Endres, M, Babnigg, G, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-10-03 | | Release date: | 2014-10-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and evolutionary relationships of "AT-less" type I polyketide synthase ketosynthases.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4WLI
 
 | | Stationary Phase Survival Protein YuiC from B.subtilis | | Descriptor: | 1,2-ETHANEDIOL, YuiC | | Authors: | Quay, D.H.X, Cole, A.R, Cryar, A, Thalassinos, K, Williams, M.A, Bhakta, S, Keep, N.H. | | Deposit date: | 2014-10-07 | | Release date: | 2015-07-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structure of the stationary phase survival protein YuiC from B.subtilis.
Bmc Struct.Biol., 15, 2015
|
|
5KPL
 
 | | Glycogen Synthase Kinase 3 beta Complexed with BRD0705 | | Descriptor: | (4~{S})-4-ethyl-7,7-dimethyl-4-phenyl-2,6,8,9-tetrahydropyrazolo[3,4-b]quinolin-5-one, Glycogen synthase kinase-3 beta | | Authors: | Lakshminarasimhan, D, White, A, Nadupalli, A, Suto, R.K. | | Deposit date: | 2016-07-04 | | Release date: | 2018-03-14 | | Last modified: | 2018-03-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Exploiting an Asp-Glu "switch" in glycogen synthase kinase 3 to design paralog-selective inhibitors for use in acute myeloid leukemia.
Sci Transl Med, 10, 2018
|
|
3D5Y
 
 | | High resolution crystal structure of 1,5-alpha-arabinanase catalytic mutant (AbnBE201A) | | Descriptor: | CALCIUM ION, Intracellular arabinanase | | Authors: | Alhassid, A, Ben David, A, Shoham, Y, Shoham, G. | | Deposit date: | 2008-05-18 | | Release date: | 2009-04-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Crystal structure of an inverting GH 43 1,5-alpha-L-arabinanase from Geobacillus stearothermophilus complexed with its substrate
Biochem.J., 422, 2009
|
|
4WNV
 
 | | Human Cytochrome P450 2D6 Quinine Complex | | Descriptor: | Cytochrome P450 2D6, GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Wang, A, Stout, C.D, Johnson, E.F. | | Deposit date: | 2014-10-14 | | Release date: | 2015-01-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Contributions of Ionic Interactions and Protein Dynamics to Cytochrome P450 2D6 (CYP2D6) Substrate and Inhibitor Binding.
J.Biol.Chem., 290, 2015
|
|
4WP4
 
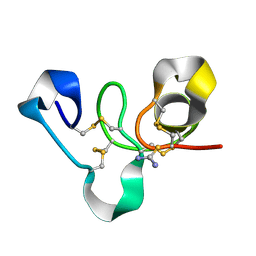 | | Hev b 6.02 (hevein) extracted from surgical gloves | | Descriptor: | Pro-hevein, THIOUREA | | Authors: | Galicia, C, Rodriguez-Romero, A. | | Deposit date: | 2014-10-17 | | Release date: | 2015-03-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Impact of the vulcanization process on the structural characteristics and IgE recognition of two allergens, Hev b 2 and Hev b 6.02, extracted from latex surgical gloves.
Mol.Immunol., 65, 2015
|
|
4X8V
 
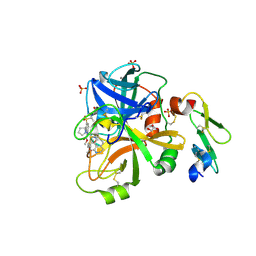 | | FACTOR VIIA IN COMPLEX WITH THE INHIBITOR (methyl {3-[(2R)-1-{(2R)-2-(3,4-dimethoxyphenyl)-2-[(1-oxo-1,2,3,4-tetrahydroisoquinolin-7-yl)amino]acetyl}pyrrolidin-2-yl]-4-(propan-2-ylsulfonyl)phenyl}carbamate) | | Descriptor: | CALCIUM ION, Factor VIIa (Heavy Chain), Factor VIIa (Light Chain), ... | | Authors: | Wei, A. | | Deposit date: | 2014-12-10 | | Release date: | 2015-03-25 | | Last modified: | 2015-04-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of Novel P1 Groups for Coagulation Factor VIIa Inhibition Using Fragment-Based Screening.
J.Med.Chem., 58, 2015
|
|
4WJL
 
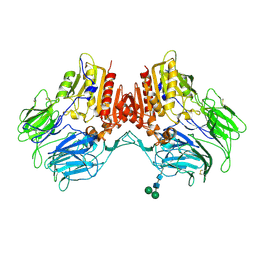 | | Structure of human dipeptidyl peptidase 10 (DPPY): a modulator of neuronal Kv4 channels | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Inactive dipeptidyl peptidase 10, ... | | Authors: | Bezerra, G.A, Dobrovetsky, E, Seitova, A, Fedosyuk, S, Dhe-Paganon, S, Gruber, K. | | Deposit date: | 2014-09-30 | | Release date: | 2015-03-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure of human dipeptidyl peptidase 10 (DPPY): a modulator of neuronal Kv4 channels.
Sci Rep, 5, 2015
|
|
4WRX
 
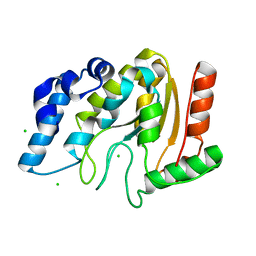 | | Crystal structure of Mycobacterium tuberculosis uracil-DNA glycosylase, Form V | | Descriptor: | CHLORIDE ION, Uracil-DNA glycosylase | | Authors: | Arif, S.M, Geethanandan, K, Mishra, P, Surolia, A, Varshney, U, Vijayan, M. | | Deposit date: | 2014-10-25 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural plasticity in Mycobacterium tuberculosis uracil-DNA glycosylase (MtUng) and its functional implications.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4WMZ
 
 | | S. cerevisiae CYP51 complexed with fluconazole in the active site | | Descriptor: | 2-(2,4-DIFLUOROPHENYL)-1,3-DI(1H-1,2,4-TRIAZOL-1-YL)PROPAN-2-OL, Lanosterol 14-alpha demethylase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sagatova, A, Keniya, M.V, Wilson, R, Wilbanks, S.M, Monk, B.C, Tyndall, J.D.A. | | Deposit date: | 2014-10-09 | | Release date: | 2015-07-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Insights into Binding of the Antifungal Drug Fluconazole to Saccharomyces cerevisiae Lanosterol 14 alpha-Demethylase.
Antimicrob.Agents Chemother., 59, 2015
|
|
4WS0
 
 | | Crystal structure of Mycobacterium tuberculosis uracil-DNA glycosylase in complex with 5-fluorouracil (A), Form II | | Descriptor: | 1,2-ETHANEDIOL, 5-FLUOROURACIL, CHLORIDE ION, ... | | Authors: | Arif, S.M, Geethanandan, K, Mishra, P, Surolia, A, Varshney, U, Vijayan, M. | | Deposit date: | 2014-10-25 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.974 Å) | | Cite: | Structural plasticity in Mycobacterium tuberculosis uracil-DNA glycosylase (MtUng) and its functional implications.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4WS3
 
 | | Crystal structure of Mycobacterium tuberculosis uracil-DNA glycosylase in complex with 6-aminouracil, Form IV | | Descriptor: | 6-aminopyrimidine-2,4(3H,5H)-dione, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Arif, S.M, Geethanandan, K, Mishra, P, Surolia, A, Varshney, U, Vijayan, M. | | Deposit date: | 2014-10-25 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural plasticity in Mycobacterium tuberculosis uracil-DNA glycosylase (MtUng) and its functional implications.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4WS8
 
 | | Crystal structure of Mycobacterium tuberculosis uracil-DNA glycosylase in complex with 2-thiouracil, Form V | | Descriptor: | 2-thioxo-2,3-dihydropyrimidin-4(1H)-one, CHLORIDE ION, Uracil-DNA glycosylase | | Authors: | Arif, S.M, Geethanandan, K, Mishra, P, Surolia, A, Varshney, U, Vijayan, M. | | Deposit date: | 2014-10-25 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural plasticity in Mycobacterium tuberculosis uracil-DNA glycosylase (MtUng) and its functional implications.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
