6JMQ
 
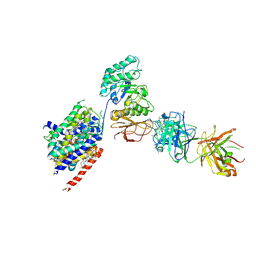 | | LAT1-CD98hc complex bound to MEM-108 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4F2 cell-surface antigen heavy chain, ... | | Authors: | Lee, Y, Nishizawa, T, Kusakizako, T, Oda, K, Ishitani, R, Nakane, T, Nureki, O. | | Deposit date: | 2019-03-13 | | Release date: | 2019-06-19 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Cryo-EM structure of the human L-type amino acid transporter 1 in complex with glycoprotein CD98hc.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6JKD
 
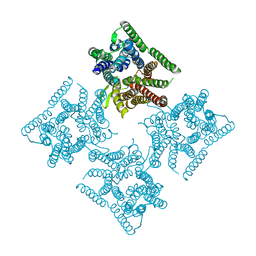 | | Crystal structure of tetrameric PepTSo2 in I4 space group | | Descriptor: | Proton:oligopeptide symporter POT family | | Authors: | Nagamura, R, Fukuda, M, Ishitani, R, Nureki, O. | | Deposit date: | 2019-02-28 | | Release date: | 2019-05-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Structural basis for oligomerization of the prokaryotic peptide transporter PepTSo2.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
6JMR
 
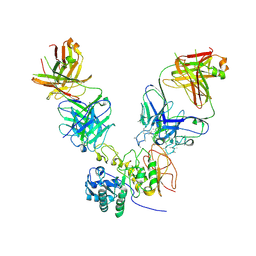 | | CD98hc extracellular domain bound to HBJ127 Fab and MEM-108 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4F2 cell-surface antigen heavy chain, ... | | Authors: | Lee, Y, Nishizawa, T, Kusakizako, T, Oda, K, Ishitani, R, Yokoyama, T, Nakane, T, Shirouzu, M, Nureki, O. | | Deposit date: | 2019-03-13 | | Release date: | 2019-06-19 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structure of the human L-type amino acid transporter 1 in complex with glycoprotein CD98hc.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6IDR
 
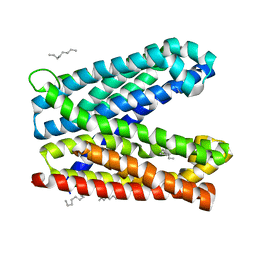 | | Crystal structure of Vibrio cholerae MATE transporter VcmN in the bent form | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, MATE family efflux transporter | | Authors: | Kusakizako, T, Claxton, D.P, Tanaka, Y, Maturana, A.D, Kuroda, T, Ishitani, R, Mchaourab, H.S, Nureki, O. | | Deposit date: | 2018-09-11 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Structural Basis of H+-Dependent Conformational Change in a Bacterial MATE Transporter.
Structure, 27, 2019
|
|
6IRY
 
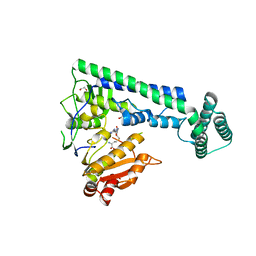 | | Crystal structure of the zebrafish cap-specific adenosine methyltransferase bound to SAH | | Descriptor: | 1,2-ETHANEDIOL, PDX1 C-terminal-inhibiting factor 1, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Hirano, S, Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2018-11-14 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cap-specific terminal N 6 -methylation of RNA by an RNA polymerase II-associated methyltransferase.
Science, 363, 2019
|
|
6JKC
 
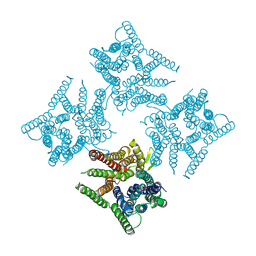 | | Crystal structure of tetrameric PepTSo2 in P4212 space group | | Descriptor: | Proton:oligopeptide symporter POT family | | Authors: | Nagamura, R, Fukuda, M, Ishitani, R, Nureki, O. | | Deposit date: | 2019-02-28 | | Release date: | 2019-05-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis for oligomerization of the prokaryotic peptide transporter PepTSo2.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
6K7H
 
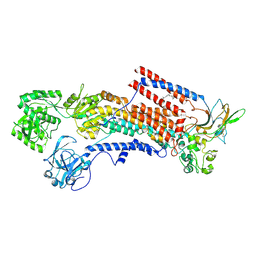 | | Cryo-EM structure of the human P4-type flippase ATP8A1-CDC50 (E1 state class2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, Cell cycle control protein 50A, ... | | Authors: | Hiraizumi, M, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2019-06-07 | | Release date: | 2019-08-28 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Cryo-EM structures capture the transport cycle of the P4-ATPase flippase.
Science, 365, 2019
|
|
6K7I
 
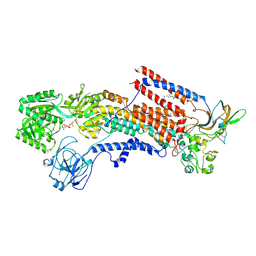 | | Cryo-EM structure of the human P4-type flippase ATP8A1-CDC50 (E1-ATP state class2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, Cell cycle control protein 50A, ... | | Authors: | Hiraizumi, M, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2019-06-07 | | Release date: | 2019-08-28 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Cryo-EM structures capture the transport cycle of the P4-ATPase flippase.
Science, 365, 2019
|
|
6K7N
 
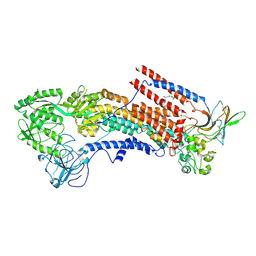 | | Cryo-EM structure of the human P4-type flippase ATP8A1-CDC50 (E1P state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, Cell cycle control protein 50A, ... | | Authors: | Hiraizumi, M, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2019-06-07 | | Release date: | 2019-08-28 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Cryo-EM structures capture the transport cycle of the P4-ATPase flippase.
Science, 365, 2019
|
|
6K7J
 
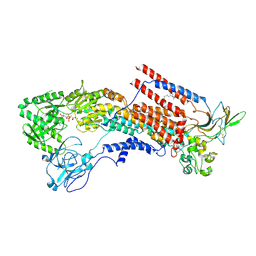 | | Cryo-EM structure of the human P4-type flippase ATP8A1-CDC50 (E1-ATP state class1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, Cell cycle control protein 50A, ... | | Authors: | Hiraizumi, M, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2019-06-07 | | Release date: | 2019-08-28 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Cryo-EM structures capture the transport cycle of the P4-ATPase flippase.
Science, 365, 2019
|
|
6K4J
 
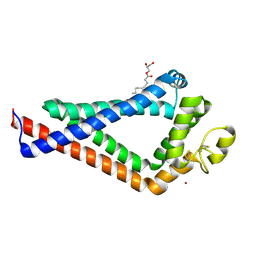 | | Crystal Structure of the the CD9 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CD9 antigen, NICKEL (II) ION, ... | | Authors: | Umeda, R, Nishizawa, T, Sato, K, Nureki, O. | | Deposit date: | 2019-05-24 | | Release date: | 2020-05-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Structural insights into tetraspanin CD9 function.
Nat Commun, 11, 2020
|
|
3VUW
 
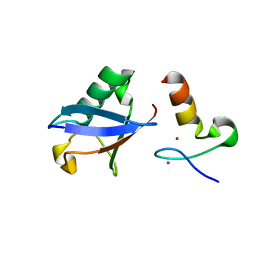 | | Crystal structure of A20 ZF7 in complex with linear ubiquitin, form I | | Descriptor: | POTASSIUM ION, Polyubiquitin-C, Tumor necrosis factor alpha-induced protein 3, ... | | Authors: | Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2012-07-09 | | Release date: | 2013-02-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Specific recognition of linear polyubiquitin by A20 zinc finger 7 is involved in NF-kappaB regulation
Embo J., 31, 2012
|
|
3VVR
 
 | | Crystal structure of MATE in complex with MaD5 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Putative uncharacterized protein, macrocyclic peptide | | Authors: | Tanaka, Y, Ishitani, R, Nureki, O. | | Deposit date: | 2012-07-27 | | Release date: | 2013-04-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for the drug extrusion mechanism by a MATE multidrug transporter.
Nature, 496, 2013
|
|
3W1W
 
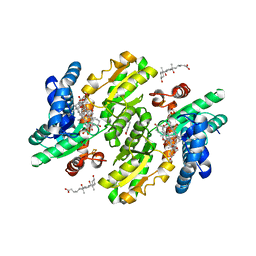 | | Protein-drug complex | | Descriptor: | 1,2-ETHANEDIOL, 2-HYDROXYBENZOIC ACID, CHOLIC ACID, ... | | Authors: | Ishii, R, Gupta, V, Yamaguchi, Y, Handa, H, Nureki, O. | | Deposit date: | 2012-11-21 | | Release date: | 2013-10-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.006 Å) | | Cite: | Salicylic Acid induces mitochondrial injury by inhibiting ferrochelatase heme biosynthesis activity
Mol.Pharmacol., 84, 2013
|
|
3VMF
 
 | | Archaeal protein | | Descriptor: | Elongation factor 1-alpha, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Kobayashi, K, Saito, K, Ishitani, R, Ito, K, Nureki, O. | | Deposit date: | 2011-12-12 | | Release date: | 2012-07-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for translation termination by archaeal RF1 and GTP-bound EF1alpha complex
Nucleic Acids Res., 40, 2012
|
|
3VWA
 
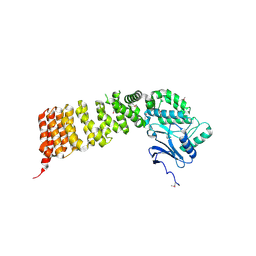 | | Crystal structure of Cex1p | | Descriptor: | Cytoplasmic export protein 1 | | Authors: | Nozawa, K, Ishitani, R, Nureki, O. | | Deposit date: | 2012-08-13 | | Release date: | 2013-06-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Cex1p reveals the mechanism of tRNA trafficking between nucleus and cytoplasm
Nucleic Acids Res., 41, 2013
|
|
3VVS
 
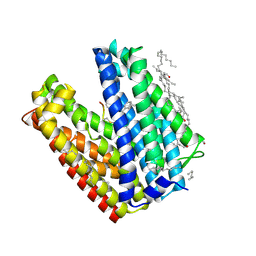 | | Crystal structure of MATE in complex with MaD3S | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Putative uncharacterized protein, macrocyclic peptide | | Authors: | Tanaka, Y, Ishitani, R, Nureki, O. | | Deposit date: | 2012-07-27 | | Release date: | 2013-04-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for the drug extrusion mechanism by a MATE multidrug transporter.
Nature, 496, 2013
|
|
3VVO
 
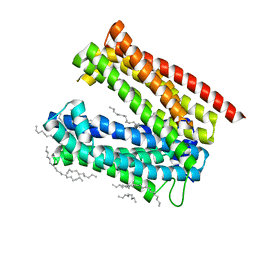 | |
3WTR
 
 | | Crystal structure of E. coli YfcM bound to Co(II) | | Descriptor: | COBALT (II) ION, Uncharacterized protein | | Authors: | Kobayashi, K, Ishii, R, Ishitani, R, Nureki, O. | | Deposit date: | 2014-04-19 | | Release date: | 2015-04-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | The non-canonical hydroxylase structure of YfcM reveals a metal ion-coordination motif required for EF-P hydroxylation
Nucleic Acids Res., 42, 2014
|
|
3WO7
 
 | | Crystal structure of YidC from Bacillus halodurans (form II) | | Descriptor: | COPPER (II) ION, Membrane protein insertase YidC 2 | | Authors: | Kumazaki, K, Tsukazaki, T, Ishitani, R, Nureki, O. | | Deposit date: | 2013-12-20 | | Release date: | 2014-04-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.201 Å) | | Cite: | Structural basis of Sec-independent membrane protein insertion by YidC.
Nature, 509, 2014
|
|
3UFZ
 
 | | Crystal structure of a Trp-less green fluorescent protein translated by the universal genetic code | | Descriptor: | Green fluorescent protein | | Authors: | Kawahara-Kobayashi, A, Araiso, Y, Matsuda, T, Yokoyama, S, Kigawa, T, Nureki, O, Kiga, D. | | Deposit date: | 2011-11-02 | | Release date: | 2012-10-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Simplification of the genetic code: restricted diversity of genetically encoded amino acids.
Nucleic Acids Res., 40, 2012
|
|
3VVP
 
 | | Crystal structure of MATE in complex with Br-NRF | | Descriptor: | 6-bromo-1-ethyl-4-oxo-7-(piperazin-1-yl)-1,4-dihydroquinoline-3-carboxylic acid, Putative uncharacterized protein | | Authors: | Tanaka, Y, Ishitani, R, Nureki, O. | | Deposit date: | 2012-07-27 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Structural basis for the drug extrusion mechanism by a MATE multidrug transporter.
Nature, 496, 2013
|
|
3WUH
 
 | | Qri7 and AMP complex | | Descriptor: | ADENOSINE MONOPHOSPHATE, ZINC ION, tRNA N6-adenosine threonylcarbamoyltransferase, ... | | Authors: | Tominaga, T, Kobayashi, K, Ishii, R, Ishitani, R, Nureki, O. | | Deposit date: | 2014-04-24 | | Release date: | 2014-09-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.937 Å) | | Cite: | Structure of Saccharomyces cerevisiae mitochondrial Qri7 in complex with AMP
ACTA CRYSTALLOGR.,SECT.F, 70, 2014
|
|
3W4T
 
 | | Crystal structure of MATE P26A mutant | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Putative uncharacterized protein | | Authors: | Tanaka, Y, Ishitani, R, Nureki, O. | | Deposit date: | 2013-01-16 | | Release date: | 2013-04-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.096 Å) | | Cite: | Structural basis for the drug extrusion mechanism by a MATE multidrug transporter.
Nature, 496, 2013
|
|
3WAX
 
 | | Crystal Structure of Autotaxin in Complex with 3BoA | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2013-05-09 | | Release date: | 2013-07-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | Screening and X-ray Crystal Structure-based Optimization of Autotaxin (ENPP2) Inhibitors, Using a Newly Developed Fluorescence Probe
Acs Chem.Biol., 8, 2013
|
|
