6WKZ
 
 | | Cocomplex structure of Deoxyhypusine synthase with inhibitor 6-[(1R)-2-AMINO-1-PHENYLETHYL]-3-(PYRIDIN-3-YL)-4H,5H,6H,7H-THIENO[2,3-C]PYRIDIN-7-ONE | | Descriptor: | 6-[(1R)-2-amino-1-phenylethyl]-3-(pyridin-3-yl)-5,6-dihydrothieno[2,3-c]pyridin-7(4H)-one, Deoxyhypusine synthase | | Authors: | Klein, M.G, Ambrus-Aikelin, G. | | Deposit date: | 2020-04-17 | | Release date: | 2020-08-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | New Series of Potent Allosteric Inhibitors of Deoxyhypusine Synthase.
Acs Med.Chem.Lett., 11, 2020
|
|
5AXQ
 
 | | Crystal structure of the catalytic domain of PDE10A complexed with highly potent and brain-penetrant PDE10A Inhibitor with 2-oxindole scaffold | | Descriptor: | 1-(cyclopropylmethyl)-4-fluoranyl-5-[5-methoxy-4-oxidanylidene-3-(2-phenylpyrazol-3-yl)pyridazin-1-yl]-3,3-dimethyl-indol-2-one, 3-[3-fluoranyl-4-[5-methoxy-4-oxidanylidene-3-(2-phenylpyrazol-3-yl)pyridazin-1-yl]phenyl]-1,3-oxazolidin-2-one, MAGNESIUM ION, ... | | Authors: | Oki, H, Zama, Y. | | Deposit date: | 2015-07-31 | | Release date: | 2015-11-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Design and synthesis of a novel 2-oxindole scaffold as a highly potent and brain-penetrant phosphodiesterase 10A inhibitor
Bioorg.Med.Chem., 23, 2015
|
|
5AXP
 
 | | Crystal structure of the catalytic domain of PDE10A complexed with 1-(2-fluoro-4-(2-oxo-1,3-oxazolidin-3-yl)phenyl)-5-methoxy-3-(1-phenyl-1H-pyrazol-5-yl)pyridazin-4(1H)-one | | Descriptor: | 3-[3-fluoranyl-4-[5-methoxy-4-oxidanylidene-3-(2-phenylpyrazol-3-yl)pyridazin-1-yl]phenyl]-1,3-oxazolidin-2-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Oki, H, Zama, Y. | | Deposit date: | 2015-07-31 | | Release date: | 2015-11-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Design and synthesis of a novel 2-oxindole scaffold as a highly potent and brain-penetrant phosphodiesterase 10A inhibitor
Bioorg.Med.Chem., 23, 2015
|
|
5XM0
 
 | | The mouse nucleosome structure containing H2A, H2B type3-A, H3.3, and H4 | | Descriptor: | DNA (146-MER), Histone H2A type 1-B, Histone H2B type 3-A, ... | | Authors: | Taguchi, H, Horikoshi, N, Kurumizaka, H. | | Deposit date: | 2017-05-12 | | Release date: | 2018-03-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.874 Å) | | Cite: | Histone H3.3 sub-variant H3mm7 is required for normal skeletal muscle regeneration.
Nat Commun, 9, 2018
|
|
5Y0D
 
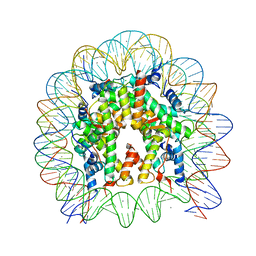 | | Crystal Structure of the human nucleosome containing the H2B E76K mutant | | Descriptor: | CHLORIDE ION, DNA (146-MER), Histone H2A type 1-B/E, ... | | Authors: | Kurumizaka, H, Arimura, Y, Fujita, R, Noda, M. | | Deposit date: | 2017-07-16 | | Release date: | 2018-07-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Cancer-associated mutations of histones H2B, H3.1 and H2A.Z.1 affect the structure and stability of the nucleosome.
Nucleic Acids Res., 46, 2018
|
|
4ZXE
 
 | | X-ray crystal structure of chitosan-binding module 1 derived from chitosanase/glucanase from Paenibacillus sp. IK-5. | | Descriptor: | 1,2-ETHANEDIOL, Glucanase/Chitosanase, SULFATE ION | | Authors: | Shinya, S, Oi, H, Kitaoku, Y, Ohnuma, T, Numata, T, Fukamizo, T. | | Deposit date: | 2015-05-20 | | Release date: | 2016-04-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Mechanism of chitosan recognition by CBM32 carbohydrate-binding modules from a Paenibacillus sp. IK-5 chitosanase/glucanase
Biochem.J., 473, 2016
|
|
4ZY9
 
 | | X-ray crystal structure of selenomethionine-labelled V110M mutant of chitosan-binding module 1 derived from chitosanase/glucanase from Paenibacillus sp. IK-5 | | Descriptor: | Glucanase/chitosanase | | Authors: | Shinya, S, Oi, H, Kitaoku, Y, Ohnuma, T, Numata, T, Fukamizo, T. | | Deposit date: | 2015-05-21 | | Release date: | 2016-04-13 | | Last modified: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Mechanism of chitosan recognition by CBM32 carbohydrate-binding modules from a Paenibacillus sp. IK-5 chitosanase/glucanase
Biochem.J., 473, 2016
|
|
5Z30
 
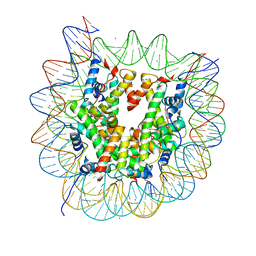 | | The crystal structure of the nucleosome containing a cancer-associated histone H2A.Z R80C mutant | | Descriptor: | CHLORIDE ION, DNA (146-MER), Histone H2A.Z, ... | | Authors: | Horikoshi, N, Arimura, Y, Kurumizaka, H. | | Deposit date: | 2018-01-05 | | Release date: | 2018-07-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Cancer-associated mutations of histones H2B, H3.1 and H2A.Z.1 affect the structure and stability of the nucleosome.
Nucleic Acids Res., 46, 2018
|
|
4ZZ5
 
 | | X-ray crystal structure of chitosan-binding module 2 derived from chitosanase/glucanase from Paenibacillus sp. IK-5 | | Descriptor: | 1,2-ETHANEDIOL, Glucanase/chitosanase, SULFATE ION | | Authors: | Shinya, S, Oi, H, Kitaoku, Y, Ohnuma, T, Numata, T, Fukamizo, T. | | Deposit date: | 2015-05-22 | | Release date: | 2016-04-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Mechanism of chitosan recognition by CBM32 carbohydrate-binding modules from a Paenibacillus sp. IK-5 chitosanase/glucanase
Biochem.J., 473, 2016
|
|
4ZZ8
 
 | | X-ray crystal structure of chitosan-binding module 2 in complex with chitotriose derived from chitosanase/glucanase from Paenibacillus sp. IK-5 | | Descriptor: | 1,2-ETHANEDIOL, 2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose, Glucanase/chitosanase, ... | | Authors: | Shinya, S, Oi, H, Kitaoku, Y, Ohnuma, T, Numata, T, Fukamizo, T. | | Deposit date: | 2015-05-22 | | Release date: | 2016-04-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Mechanism of chitosan recognition by CBM32 carbohydrate-binding modules from a Paenibacillus sp. IK-5 chitosanase/glucanase
Biochem.J., 473, 2016
|
|
5Y0C
 
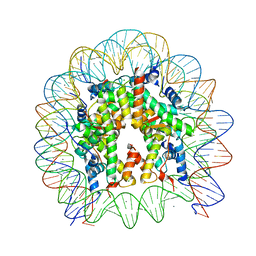 | | Crystal Structure of the human nucleosome at 2.09 angstrom resolution | | Descriptor: | CHLORIDE ION, DNA (146-MER), Histone H2A type 1-B/E, ... | | Authors: | Kurumizaka, H, Arimura, Y, Fujita, R, Noda, M. | | Deposit date: | 2017-07-16 | | Release date: | 2018-07-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.087 Å) | | Cite: | Cancer-associated mutations of histones H2B, H3.1 and H2A.Z.1 affect the structure and stability of the nucleosome.
Nucleic Acids Res., 46, 2018
|
|
6KXV
 
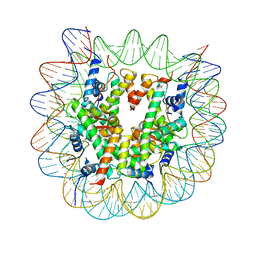 | | Crystal structure of a nucleosome containing Leishmania histone H3 | | Descriptor: | DNA (146-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Dacher, M, Taguchi, H, Kujirai, T, Kurumizaka, H. | | Deposit date: | 2019-09-13 | | Release date: | 2020-07-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.63 Å) | | Cite: | Incorporation and influence of Leishmania histone H3 in chromatin.
Nucleic Acids Res., 47, 2019
|
|
5YBG
 
 | | Crystal structure of the GluA2o LBD in complex with glutamate and LY451395 | | Descriptor: | ACETATE ION, GLUTAMIC ACID, GLYCEROL, ... | | Authors: | Sogabe, S, Igaki, S, Hirokawa, A, Zama, Y, Lane, W, Snell, G. | | Deposit date: | 2017-09-04 | | Release date: | 2018-01-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | HBT1, a Novel AMPA Receptor Potentiator with Lower Agonistic Effect, Avoided Bell-Shaped Response in In Vitro BDNF Production.
J. Pharmacol. Exp. Ther., 364, 2018
|
|
5ZG3
 
 | | Crystal structure of the GluA2o LBD in complex with glutamate and TAK-137 | | Descriptor: | 9-(4-phenoxyphenyl)-3,4-dihydro-2H-2lambda~6~-pyrido[2,1-c][1,2,4]thiadiazine-2,2-dione, ACETATE ION, GLUTAMIC ACID, ... | | Authors: | Sogabe, S, Igaki, S, Hirokawa, A, Zama, Y, Lane, W, Snell, G. | | Deposit date: | 2018-03-07 | | Release date: | 2019-01-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | TAK-137, an AMPA-R potentiator with little agonistic effect, has a wide therapeutic window.
Neuropsychopharmacology, 44, 2019
|
|
5ZG1
 
 | | Crystal structure of the GluA2o LBD in complex with glutamate and Compound-2 | | Descriptor: | 9-(4-~{tert}-butylphenyl)-3,4-dihydropyrido[2,1-c][1,2,4]thiadiazine 2,2-dioxide, GLUTAMIC ACID, GLYCEROL, ... | | Authors: | Sogabe, S, Igaki, S, Hirokawa, A, Zama, Y, Lane, W, Snell, G. | | Deposit date: | 2018-03-07 | | Release date: | 2019-01-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | TAK-137, an AMPA-R potentiator with little agonistic effect, has a wide therapeutic window.
Neuropsychopharmacology, 44, 2019
|
|
5ZG2
 
 | | Crystal structure of the GluA2o LBD in complex with ZK200775 and Compound-2 | | Descriptor: | 9-(4-~{tert}-butylphenyl)-3,4-dihydropyrido[2,1-c][1,2,4]thiadiazine 2,2-dioxide, ACETATE ION, GLYCEROL, ... | | Authors: | Sogabe, S, Igaki, S, Hirokawa, A, Zama, Y, Lane, W, Snell, G. | | Deposit date: | 2018-03-07 | | Release date: | 2019-01-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | TAK-137, an AMPA-R potentiator with little agonistic effect, has a wide therapeutic window.
Neuropsychopharmacology, 44, 2019
|
|
5ZG0
 
 | | Crystal structure of the GluA2o LBD in complex with glutamate and Compound-1 | | Descriptor: | 9-{4-[(propan-2-yl)oxy]phenyl}-3,4-dihydro-2H-2lambda~6~-pyrido[2,1-c][1,2,4]thiadiazine-2,2-dione, ACETATE ION, GLUTAMIC ACID, ... | | Authors: | Sogabe, S, Igaki, S, Hirokawa, A, Zama, Y, Lane, W, Snell, G. | | Deposit date: | 2018-03-07 | | Release date: | 2019-01-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | TAK-137, an AMPA-R potentiator with little agonistic effect, has a wide therapeutic window.
Neuropsychopharmacology, 44, 2019
|
|
5YBF
 
 | | Crystal structure of the GluA2o LBD in complex with glutamate and HBT1 | | Descriptor: | 2-[2-[5-methyl-3-(trifluoromethyl)pyrazol-1-yl]ethanoylamino]-4,5,6,7-tetrahydro-1-benzothiophene-3-carboxamide, ACETATE ION, GLUTAMIC ACID, ... | | Authors: | Sogabe, S, Igaki, S, Hirokawa, A, Zama, Y, Lane, W, Snell, G. | | Deposit date: | 2017-09-04 | | Release date: | 2018-01-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | HBT1, a Novel AMPA Receptor Potentiator with Lower Agonistic Effect, Avoided Bell-Shaped Response in In Vitro BDNF Production.
J. Pharmacol. Exp. Ther., 364, 2018
|
|
5W4B
 
 | | The crystal structure of human S-adenosylhomocysteine hydrolase (AHCY) bound to benzothiazole inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 4-[(2,5-dioxo-2,5-dihydro-1H-imidazol-1-yl)methyl]-N-[2-(morpholin-4-yl)-1,3-benzothiazol-6-yl]benzamide, Adenosylhomocysteinase, ... | | Authors: | Dougan, D.R, Lawson, J.D, Lane, W. | | Deposit date: | 2017-06-09 | | Release date: | 2017-06-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Identification of AHCY inhibitors using novel high-throughput mass spectrometry.
Biochem. Biophys. Res. Commun., 491, 2017
|
|
6JOU
 
 | | Crystal structure of the human nucleosome containing H2A.Z.1 S42R | | Descriptor: | DNA (146-MER), Histone H2A.Z, Histone H2B type 1-J, ... | | Authors: | Horikoshi, N, Sato, K, Mizukami, Y, Kurumizaka, H. | | Deposit date: | 2019-03-23 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structure-based design of an H2A.Z.1 mutant stabilizing a nucleosome in vitro and in vivo.
Biochem.Biophys.Res.Commun., 515, 2019
|
|
5W49
 
 | | The crystal structure of human S-adenosylhomocysteine hydrolase (AHCY) bound to oxadiazole inhibitor | | Descriptor: | (4-amino-1,2,5-oxadiazol-3-yl)[(3R)-3-{4-[(3-methoxyphenyl)amino]-6-methylpyridin-2-yl}pyrrolidin-1-yl]methanone, 1,2-ETHANEDIOL, Adenosylhomocysteinase, ... | | Authors: | Dougan, D.R, Lawson, J.D, Lane, W. | | Deposit date: | 2017-06-09 | | Release date: | 2017-06-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Identification of AHCY inhibitors using novel high-throughput mass spectrometry.
Biochem. Biophys. Res. Commun., 491, 2017
|
|
3VOO
 
 | | Cytochrome P450SP alpha (CYP152B1) mutant A245E | | Descriptor: | Fatty acid alpha-hydroxylase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Fujishiro, T, Shoji, O, Sugimoto, H, Shiro, Y, Watanabe, Y. | | Deposit date: | 2012-01-31 | | Release date: | 2013-02-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | A substrate-binding-state mimic of H2O2-dependent cytochrome P450 produced by one-point mutagenesis and peroxygenation of non-native substrates
Catalysis Science And Technology, 6, 2016
|
|
3VNO
 
 | | Cytochrome P450SP alpha (CYP152B1) mutant R241E | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Fatty acid alpha-hydroxylase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Fujishiro, T, Shoji, O, Sugimoto, H, Shiro, Y, Watanabe, Y. | | Deposit date: | 2012-01-17 | | Release date: | 2013-02-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | A substrate-binding-state mimic of H2O2-dependent cytochrome P450 produced by one-point mutagenesis and peroxygenation of non-native substrates
Catalysis Science And Technology, 6, 2016
|
|
3VTJ
 
 | | Cytochrome P450SP alpha (CYP152B1) mutant A245H | | Descriptor: | Fatty acid alpha-hydroxylase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Fujishiro, T, Shoji, O, Sugimoto, H, Shiro, Y, Watanabe, Y. | | Deposit date: | 2012-05-30 | | Release date: | 2013-06-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | A substrate-binding-state mimic of H2O2-dependent cytochrome P450 produced by one-point mutagenesis and peroxygenation of non-native substrates
Catalysis Science And Technology, 6, 2016
|
|
3WYM
 
 | | Crystal structure of the catalytic domain of PDE10A complexed with 1-(2-fluoro-4-(1H-pyrazol-1-yl)phenyl)-5-methoxy-3-(1-phenyl-1H-pyrazol-5-yl)pyridazin-4(1H)-one | | Descriptor: | 1-[2-fluoro-4-(1H-pyrazol-1-yl)phenyl]-5-methoxy-3-(1-phenyl-1H-pyrazol-5-yl)pyridazin-4(1H)-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Oki, H, Hayano, Y. | | Deposit date: | 2014-09-01 | | Release date: | 2014-11-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of 1-[2-fluoro-4-(1H-pyrazol-1-yl)phenyl]-5-methoxy-3-(1-phenyl-1H-pyrazol-5-yl)pyridazin-4(1H)-one (TAK-063), a highly potent, selective, and orally active phosphodiesterase 10A (PDE10A) inhibitor.
J.Med.Chem., 57, 2014
|
|
