7K6D
 
 | | SARS-CoV-2 Main Protease Co-Crystal Structure with Telaprevir Determined from Crystals Grown with 40 nL Acoustically Ejected Mpro Droplets at 1.48 A Resolution (Cryo-protected) | | Descriptor: | (1S,3aR,6aS)-2-[(2S)-2-({(2S)-2-cyclohexyl-2-[(pyrazin-2-ylcarbonyl)amino]acetyl}amino)-3,3-dimethylbutanoyl]-N-[(2R,3S)-1-(cyclopropylamino)-2-hydroxy-1-oxohexan-3-yl]octahydrocyclopenta[c]pyrrole-1-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Kreitler, D.F, Andi, B, Kumaran, D, Soares, A.S, Shi, W, Jakoncic, J, Fuchs, M.R, Keereetaweep, J, Shanklin, J, McSweeney, S. | | Deposit date: | 2020-09-19 | | Release date: | 2020-09-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Hepatitis C virus NS3/4A inhibitors and other drug-like compounds as covalent binders of SARS-CoV-2 main protease.
Sci Rep, 12, 2022
|
|
7K6E
 
 | | SARS-CoV-2 Main Protease Co-Crystal Structure with Telaprevir Determined from Crystals Grown with 40 nL Acoustically Ejected Mpro Droplets at 1.63 A Resolution (Direct Vitrification) | | Descriptor: | (1S,3aR,6aS)-2-[(2S)-2-({(2S)-2-cyclohexyl-2-[(pyrazin-2-ylcarbonyl)amino]acetyl}amino)-3,3-dimethylbutanoyl]-N-[(2R,3S)-1-(cyclopropylamino)-2-hydroxy-1-oxohexan-3-yl]octahydrocyclopenta[c]pyrrole-1-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Kreitler, D.F, Andi, B, Kumaran, D, Soares, A.S, Shi, W, Jakoncic, J, Fuchs, M.R, Keereetaweep, J, Shanklin, J, McSweeney, S. | | Deposit date: | 2020-09-19 | | Release date: | 2020-09-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Hepatitis C virus NS3/4A inhibitors and other drug-like compounds as covalent binders of SARS-CoV-2 main protease.
Sci Rep, 12, 2022
|
|
4GRN
 
 | |
4BFQ
 
 | | Assembly of a triple pi-stack of ligands in the binding site of Aplysia californica acetylcholine binding protein (AChBP) | | Descriptor: | 4,6-dimethyl-N'-(3-pyridin-2-ylisoquinolin-1-yl)pyrimidine-2-carboximidamide, GLYCEROL, SOLUBLE ACETYLCHOLINE RECEPTOR | | Authors: | Stornaiuolo, M, De Kloe, G.E, Rucktooa, P, Fish, A, van Elk, R, Edink, E.S, Bertrand, D, Smit, A.B, de Esch, I.J.P, Sixma, T.K. | | Deposit date: | 2013-03-21 | | Release date: | 2013-05-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Assembly of a Pi-Pi Stack of Ligands in the Binding Site of an Acetylcholine Binding Protein
Nat.Commun., 4, 2013
|
|
4AK8
 
 | | Structure of F241L mutant of langerin carbohydrate recognition domain. | | Descriptor: | C-TYPE LECTIN DOMAIN FAMILY 4 MEMBER K, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Chabrol, E, Thepaut, M, Dezutter-Dambuyant, C, Vives, C, Marcoux, J, Kahn, R, Valadeau-Guilemond, J, Vachette, P, Durand, D, Fieschi, F. | | Deposit date: | 2012-02-22 | | Release date: | 2013-04-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Alteration of the Langerin Oligomerization State Affects Birbeck Granule Formation.
Biophys.J., 108, 2015
|
|
4AKK
 
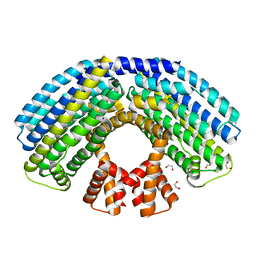 | | Structure of the NasR transcription antiterminator | | Descriptor: | 1,2-ETHANEDIOL, NITRATE REGULATORY PROTEIN | | Authors: | Boudes, M, Lazar, N, Graille, M, Durand, D, Gaidenko, T.A, Stewart, V, van Tilbeurgh, H. | | Deposit date: | 2012-02-24 | | Release date: | 2012-06-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.145 Å) | | Cite: | The Structure of the Nasr Transcription Antiterminator Reveals a One-Component System with a Nit Nitrate Receptor Coupled to an Antar RNA-Binding Effector.
Mol.Microbiol., 85, 2012
|
|
4AEA
 
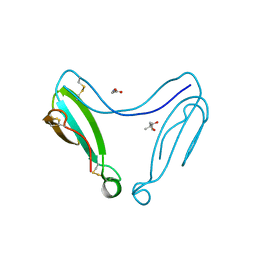 | | Dimeric alpha-cobratoxin X-ray structure: Localization of intermolecular disulfides and possible mode of binding to nicotinic acetylcholine receptors | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GLYCINE, LONG NEUROTOXIN 1 | | Authors: | Rucktooa, P, Osipov, A.V, Kasheverov, I.E, Filkin, S.Y, Starkov, V.G, Andreeva, T.V, Bertrand, D, Utkin, Y.N, Tsetlin, V.I, Sixma, T.K. | | Deposit date: | 2012-01-09 | | Release date: | 2012-01-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Dimeric Alpha-Cobratoxin X-Ray Structure: Localization of Intermolecular Disulfides and Possible Mode of Binding to Nicotinic Acetylcholine Receptors.
J.Biol.Chem., 287, 2012
|
|
4A8E
 
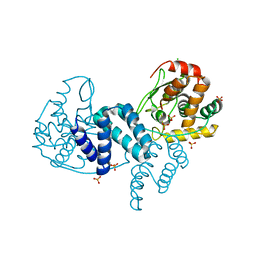 | | The structure of a dimeric Xer recombinase from archaea | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, PROBABLE TYROSINE RECOMBINASE XERC-LIKE, ... | | Authors: | Brooks, M.A, ElArnaout, T, Duranda, D, Lisboa, J, Lazar, N, Raynal, B, vanTilbeurgh, H, Serre, M, Quevillon-Cheruel, S. | | Deposit date: | 2011-11-21 | | Release date: | 2012-12-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | The Carboxy-Terminal Alpha N Helix of the Archaeal Xera Tyrosine Recombinase is a Molecular Switch to Control Site-Specific Recombination.
Plos One, 8, 2013
|
|
3ZKR
 
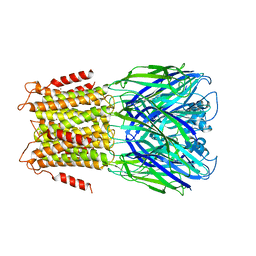 | | X-ray structure of a pentameric ligand gated ion channel from Erwinia chrysanthemi (ELIC) in complex with bromoform | | Descriptor: | CYS-LOOP LIGAND-GATED ION CHANNEL, TRIBROMOMETHANE | | Authors: | Spurny, R, Billen, B, Howard, R.J, Brams, M, Debaveye, S, Price, K.L, Weston, D.A, Strelkov, S.V, Tytgat, J, Bertrand, S, Bertrand, D, Lummis, S.C.R, Ulens, C. | | Deposit date: | 2013-01-24 | | Release date: | 2013-02-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.649 Å) | | Cite: | Multisite Binding of a General Anesthetic to the Prokaryotic Pentameric Erwinia Chrysanthemi Ligand-Gated Ion Channel (Elic).
J.Biol.Chem., 288, 2013
|
|
4CBV
 
 | | X-ray structure of full-length ComE from Streptococcus pneumoniae. | | Descriptor: | COME | | Authors: | Boudes, M, Durand, D, Graille, M, van Tilbeurgh, H, Quevillon-Cheruel, S. | | Deposit date: | 2013-10-16 | | Release date: | 2014-02-12 | | Last modified: | 2014-05-14 | | Method: | X-RAY DIFFRACTION (3.39 Å) | | Cite: | Structural Insights Into the Dimerization of the Response Regulator Come from Streptococcus Pneumoniae.
Nucleic Acids Res., 42, 2014
|
|
3KER
 
 | |
5NLV
 
 | | Brag2 Sec7-PH (390-763) | | Descriptor: | IQ motif and SEC7 domain-containing protein 1 | | Authors: | Nawrotek, A, Cherfils, J. | | Deposit date: | 2017-04-05 | | Release date: | 2017-09-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Multiple interactions between an Arf/GEF complex and charged lipids determine activation kinetics on the membrane.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5NBI
 
 | |
5NLY
 
 | | Brag2 Sec7-PH (390-763), P212121 | | Descriptor: | IQ motif and SEC7 domain-containing protein 1, PHOSPHATE ION | | Authors: | Nawrotek, A, Cherfils, J. | | Deposit date: | 2017-04-05 | | Release date: | 2017-09-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Multiple interactions between an Arf/GEF complex and charged lipids determine activation kinetics on the membrane.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5NB5
 
 | |
6MOA
 
 | | C-terminal bromodomain of human BRD2 in complex with 4-(2-cyclopropyl-7-(6-methylquinolin-5-yl)-1H-benzo[d]imidazol-5-yl)-3,5-dimethylisoxazole inhibitor | | Descriptor: | 4-(2-cyclopropyl-7-(6-methylquinolin-5-yl)-1H-benzo[d]imidazol-5-yl)-3,5-dimethylisoxazole, Bromodomain-containing protein 2, GLYCEROL | | Authors: | Lansdon, E.B, Newby, Z.E.R. | | Deposit date: | 2018-10-04 | | Release date: | 2019-01-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.271 Å) | | Cite: | Structure-guided discovery of a novel, potent, and orally bioavailable 3,5-dimethylisoxazole aryl-benzimidazole BET bromodomain inhibitor.
Bioorg. Med. Chem., 27, 2019
|
|
6MO7
 
 | | N-terminal bromodomain of human BRD2 with N-((4-(3-(N-cyclopentylsulfamoyl)-4-methylphenyl)-3-methylisoxazol-5-yl)methyl)acetamide inhibitor | | Descriptor: | Bromodomain-containing protein 2, N-({4-[3-(cyclopentylsulfamoyl)-4-methylphenyl]-3-methyl-1,2-oxazol-5-yl}methyl)acetamide | | Authors: | Lansdon, E.B, Newby, Z.E.R. | | Deposit date: | 2018-10-04 | | Release date: | 2019-01-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-guided discovery of a novel, potent, and orally bioavailable 3,5-dimethylisoxazole aryl-benzimidazole BET bromodomain inhibitor.
Bioorg. Med. Chem., 27, 2019
|
|
6MO9
 
 | | N-terminal bromodomain of human BRD2 in complex with N-cyclopentyl-7-(3,5-dimethylisoxazol-4-yl)quinoline-5-sulfonamide inhibitor | | Descriptor: | Bromodomain-containing protein 2, N-cyclopentyl-7-(3,5-dimethyl-1,2-oxazol-4-yl)quinoline-5-sulfonamide | | Authors: | Lansdon, E.B, Newby, Z.E.R. | | Deposit date: | 2018-10-04 | | Release date: | 2019-01-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structure-guided discovery of a novel, potent, and orally bioavailable 3,5-dimethylisoxazole aryl-benzimidazole BET bromodomain inhibitor.
Bioorg. Med. Chem., 27, 2019
|
|
6MO8
 
 | | N-terminal bromodomain of human BRD2 in complex with 4,4'-(quinoline-5,7-diyl)bis(3,5-dimethylisoxazole) inhibitor | | Descriptor: | 5,7-bis(3,5-dimethyl-1,2-oxazol-4-yl)quinoline, Bromodomain-containing protein 2, SULFATE ION | | Authors: | Lansdon, E.B, Newby, Z.E.R. | | Deposit date: | 2018-10-04 | | Release date: | 2019-01-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-guided discovery of a novel, potent, and orally bioavailable 3,5-dimethylisoxazole aryl-benzimidazole BET bromodomain inhibitor.
Bioorg. Med. Chem., 27, 2019
|
|
3KAN
 
 | |
1I4X
 
 | |
7Z06
 
 | |
1GGQ
 
 | |
7KP6
 
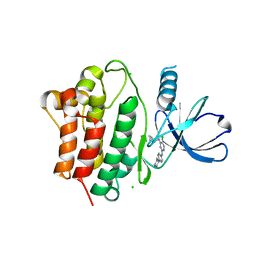 | | Structure of Ack1 kinase in complex with a selective inhibitor | | Descriptor: | 5-chloro-N~2~-[4-(4-methylpiperazin-1-yl)phenyl]-N~4~-{[(2R)-oxolan-2-yl]methyl}pyrimidine-2,4-diamine, Activated CDC42 kinase 1, CHLORIDE ION | | Authors: | Thakur, M.K, Miller, W.T, Mahajan, N, Seeliger, M.A. | | Deposit date: | 2020-11-10 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Inhibiting ACK1-mediated phosphorylation of C-terminal Src kinase counteracts prostate cancer immune checkpoint blockade resistance.
Nat Commun, 13, 2022
|
|
1JZT
 
 | |
