5DET
 
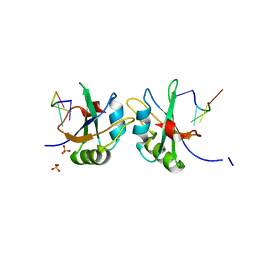 | | X-ray structure of human RBPMS in complex with the RNA | | Descriptor: | RNA (5'-R(*UP*CP*AP*C)-3'), RNA (5'-R(P*UP*CP*AP*CP*U)-3'), RNA-binding protein with multiple splicing, ... | | Authors: | Teplova, M, Farazi, T.A, Tuschl, T, Patel, D.J. | | Deposit date: | 2015-08-25 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis underlying CAC RNA recognition by the RRM domain of dimeric RNA-binding protein RBPMS.
Q. Rev. Biophys., 49, 2016
|
|
6PPU
 
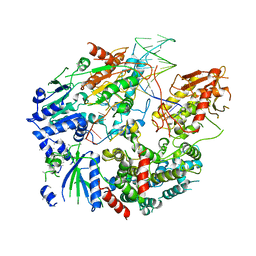 | | Cryo-EM structure of AdnAB-AMPPNP-DNA complex | | Descriptor: | ATP-dependent DNA helicase (UvrD/REP), DNA (29-MER), IRON/SULFUR CLUSTER, ... | | Authors: | Jia, N, Unciuleac, M, Shuman, S, Patel, D.J. | | Deposit date: | 2019-07-08 | | Release date: | 2019-11-20 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structures and single-molecule analysis of bacterial motor nuclease AdnAB illuminate the mechanism of DNA double-strand break resection.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
201D
 
 | |
5DAH
 
 | |
202D
 
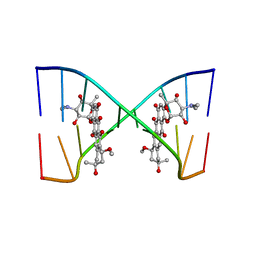 | |
2ASL
 
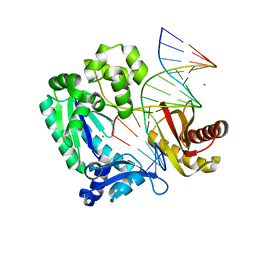 | | oxoG-modified Postinsertion Binary Complex | | Descriptor: | 5'-D(*CP*T*AP*AP*CP*(8OG)P*CP*TP*AP*CP*CP*AP*TP*CP*CP*AP*AP*CP*C)-3', 5'-D(*GP*GP*TP*TP*GP*GP*AP*TP*GP*GP*TP*AP*GP*(DOC))-3', CALCIUM ION, ... | | Authors: | Rechkoblit, O, Malinina, L, Cheng, Y, Kuryavyi, V, Broyde, S, Geacintov, N.E, Patel, D.J. | | Deposit date: | 2005-08-23 | | Release date: | 2006-01-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Stepwise Translocation of Dpo4 Polymerase during Error-Free Bypass of an oxoG Lesion
Plos Biol., 4, 2006
|
|
3O7V
 
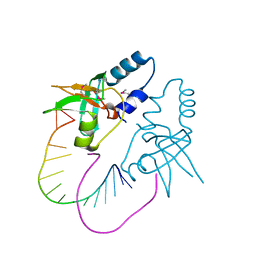 | | Crystal Structure of human Hiwi1 (V361M) PAZ domain (residues 277-399) in complex with 14-mer RNA (12-bp + 2-nt overhang) containing 2'-OCH3 at its 3'-end | | Descriptor: | Piwi-like protein 1, RNA (5'-R(*GP*CP*GP*AP*AP*UP*AP*UP*UP*CP*GP*CP*UP*(OMU))-3') | | Authors: | Tian, Y, Simanshu, D.K, Ma, J.-B, Patel, D.J. | | Deposit date: | 2010-08-01 | | Release date: | 2011-01-12 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Inaugural Article: Structural basis for piRNA 2'-O-methylated 3'-end recognition by Piwi PAZ (Piwi/Argonaute/Zwille) domains.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
5DEA
 
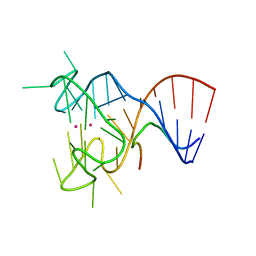 | | Crystal structure of the complex between human FMRP RGG motif and G-quadruplex RNA, cesium bound form. | | Descriptor: | CESIUM ION, Fragile X mental retardation protein 1, POTASSIUM ION, ... | | Authors: | Vasilyev, N, Polonskaia, A, Darnell, J.C, Darnell, R.B, Patel, D.J, Serganov, A. | | Deposit date: | 2015-08-25 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7973 Å) | | Cite: | Crystal structure reveals specific recognition of a G-quadruplex RNA by a beta-turn in the RGG motif of FMRP.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2ANR
 
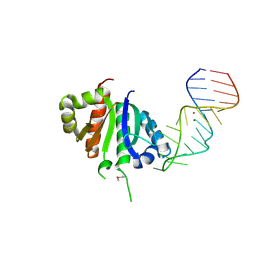 | | Crystal structure (II) of Nova-1 KH1/KH2 domain tandem with 25nt RNA hairpin | | Descriptor: | 5'-R(*CP*(5BU)P*CP*GP*CP*GP*GP*AP*UP*CP*AP*GP*UP*CP*AP*CP*CP*CP*AP*AP*GP*CP*GP*AP*G)-3', MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Malinina, L, Teplova, M, Musunuru, K, Teplov, A, Darnell, J.C, Burley, S.K, Darnell, R.B, Patel, D.J. | | Deposit date: | 2005-08-11 | | Release date: | 2006-10-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Protein-RNA and protein-protein recognition by dual KH1/2 domains of the neuronal splicing factor Nova-1.
Structure, 19, 2011
|
|
6VRC
 
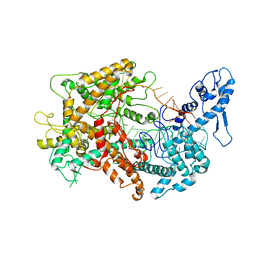 | | Cryo-EM structure of Cas13(crRNA) | | Descriptor: | CRISPR-associated endoribonuclease Cas13a, RNA (51-MER) | | Authors: | Jia, N, Meeske, A.J, Marraffini, L.A, Patel, D.J. | | Deposit date: | 2020-02-07 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A phage-encoded anti-CRISPR enables complete evasion of type VI-A CRISPR-Cas immunity.
Science, 369, 2020
|
|
3O6E
 
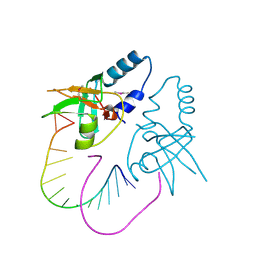 | | Crystal Structure of human Hiwi1 PAZ domain (residues 277-399) in complex with 14-mer RNA (12-bp + 2-nt overhang) containing 2'-OCH3 at its 3'-end | | Descriptor: | Piwi-like protein 1, RNA (5'-R(*GP*CP*GP*AP*AP*UP*AP*UP*UP*CP*GP*CP*UP*(OMU))-3') | | Authors: | Tian, Y, Simanshu, D.K, Ma, J.-B, Patel, D.J. | | Deposit date: | 2010-07-28 | | Release date: | 2011-01-12 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.904 Å) | | Cite: | Inaugural Article: Structural basis for piRNA 2'-O-methylated 3'-end recognition by Piwi PAZ (Piwi/Argonaute/Zwille) domains.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
5DAG
 
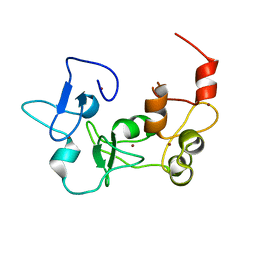 | |
5DDQ
 
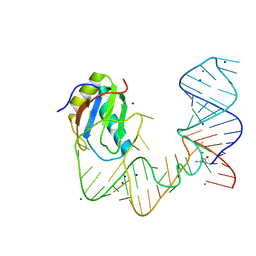 | | L-glutamine riboswitch bound with L-glutamine soaked with Mn2+ | | Descriptor: | GLUTAMINE, L-glutamine riboswitch RNA (61-MER), MAGNESIUM ION, ... | | Authors: | Ren, A, Patel, D.J. | | Deposit date: | 2015-08-25 | | Release date: | 2015-12-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Dynamic Basis for Low-Affinity, High-Selectivity Binding of L-Glutamine by the Glutamine Riboswitch.
Cell Rep, 13, 2015
|
|
207D
 
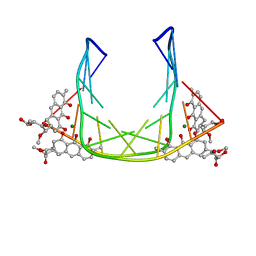 | | SOLUTION STRUCTURE OF MITHRAMYCIN DIMERS BOUND TO PARTIALLY OVERLAPPING SITES ON DNA | | Descriptor: | 1,2-HYDRO-1-OXY-3,4-HYDRO-3-(1-METHOXY-2-OXY-3,4-DIHYDROXYPENTYL)-8,9-DIHYROXY-7-METHYLANTHRACENE, 2,6-dideoxy-3-C-methyl-beta-D-ribo-hexopyranose-(1-3)-2,6-dideoxy-beta-D-galactopyranose-(1-3)-beta-D-Olivopyranose, DNA (5'-D(*TP*AP*GP*CP*TP*AP*GP*CP*TP*A)-3'), ... | | Authors: | Sastry, M, Fiala, R, Patel, D.J. | | Deposit date: | 1995-04-20 | | Release date: | 1995-09-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of mithramycin dimers bound to partially overlapping sites on DNA.
J.Mol.Biol., 251, 1995
|
|
5DDR
 
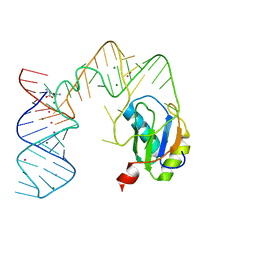 | | L-glutamine riboswitch bound with L-glutamine soaked with Cs+ | | Descriptor: | CESIUM ION, GLUTAMINE, L-glutamine riboswitch RNA (61-MER), ... | | Authors: | Ren, A, Patel, D.J. | | Deposit date: | 2015-08-25 | | Release date: | 2015-12-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.605 Å) | | Cite: | Structural and Dynamic Basis for Low-Affinity, High-Selectivity Binding of L-Glutamine by the Glutamine Riboswitch.
Cell Rep, 13, 2015
|
|
5DE8
 
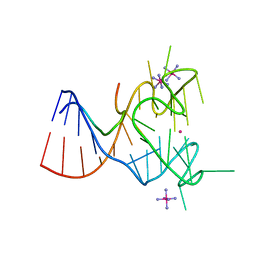 | | Crystal structure of the complex between human FMRP RGG motif and G-quadruplex RNA, iridium hexammine bound form. | | Descriptor: | Fragile X mental retardation protein 1, IRIDIUM HEXAMMINE ION, POTASSIUM ION, ... | | Authors: | Vasilyev, N, Polonskaia, A, Darnell, J.C, Darnell, R.B, Patel, D.J, Serganov, A. | | Deposit date: | 2015-08-25 | | Release date: | 2015-09-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.1003 Å) | | Cite: | Crystal structure reveals specific recognition of a G-quadruplex RNA by a beta-turn in the RGG motif of FMRP.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5CYJ
 
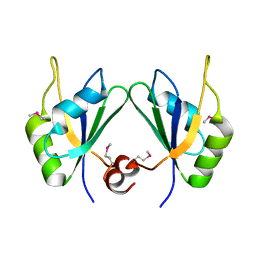 | | X-ray structure of human RBPMS | | Descriptor: | RNA-binding protein with multiple splicing | | Authors: | Teplova, M, Farazi, T.A, Patel, D.J. | | Deposit date: | 2015-07-30 | | Release date: | 2015-09-30 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural basis underlying CAC RNA recognition by the RRM domain of dimeric RNA-binding protein RBPMS.
Q. Rev. Biophys., 49, 2016
|
|
6UFK
 
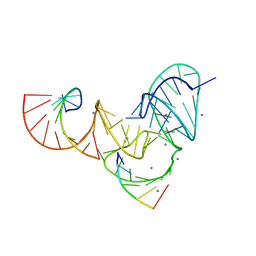 | | Pistol ribozyme product crystal soaked in Mn2+ | | Descriptor: | MANGANESE (II) ION, RNA (5'-R(*UP*CP*CP*AP*G)-3'), RNA (5'-R(*UP*CP*UP*GP*CP*UP*CP*UP*CP*(23G))-3'), ... | | Authors: | Teplova, M, Falschlunger, C, Krasheninina, O, Patel, D.J, Micura, R. | | Deposit date: | 2019-09-24 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crucial Roles of Two Hydrated Mg2+Ions in Reaction Catalysis of the Pistol Ribozyme.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
3OWW
 
 | |
3OXE
 
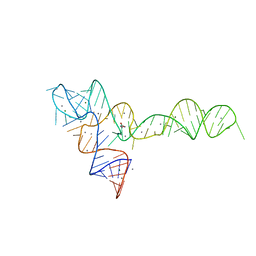 | | crystal structure of glycine riboswitch, Mn2+ soaked | | Descriptor: | GLYCINE, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Huang, L, Serganov, A, Patel, D.J. | | Deposit date: | 2010-09-21 | | Release date: | 2010-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.899 Å) | | Cite: | Structural insights into ligand recognition by a sensing domain of the cooperative glycine riboswitch.
Mol.Cell, 40, 2010
|
|
1ZH5
 
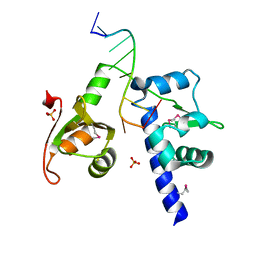 | | Structural basis for recognition of UUUOH 3'-terminii of nascent RNA pol III transcripts by La autoantigen | | Descriptor: | 5'-R(*UP*GP*CP*UP*GP*UP*UP*UP*U)-3', Lupus La protein, SULFATE ION | | Authors: | Teplova, M, Yuan, Y.R, Ilin, S, Malinina, L, Phan, A.T, Teplov, A, Patel, D.J. | | Deposit date: | 2005-04-22 | | Release date: | 2006-01-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Basis for Recognition and Sequestration of UUU(OH) 3' Temini of Nascent RNA Polymerase III Transcripts by La, a Rheumatic Disease Autoantigen.
Mol.Cell, 21, 2006
|
|
5DE5
 
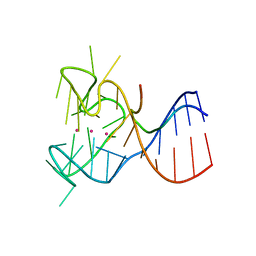 | | Crystal structure of the complex between human FMRP RGG motif and G-quadruplex RNA. | | Descriptor: | Fragile X mental retardation protein 1, POTASSIUM ION, sc1 | | Authors: | Vasilyev, N, Polonskaia, A, Darnell, J.C, Darnell, R.B, Patel, D.J, Serganov, A. | | Deposit date: | 2015-08-25 | | Release date: | 2015-09-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.0011 Å) | | Cite: | Crystal structure reveals specific recognition of a G-quadruplex RNA by a beta-turn in the RGG motif of FMRP.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3QZT
 
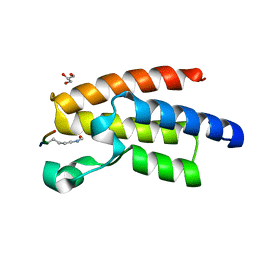 | | Crystal Structure of BPTF bromo in complex with histone H4K16ac - Form II | | Descriptor: | GLYCEROL, Histone H4, Nucleosome-remodeling factor subunit BPTF | | Authors: | Li, H, Ruthenburg, A.J, Patel, D.J. | | Deposit date: | 2011-03-07 | | Release date: | 2011-06-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Recognition of a Mononucleosomal Histone Modification Pattern by BPTF via Multivalent Interactions.
Cell(Cambridge,Mass.), 145, 2011
|
|
5IX1
 
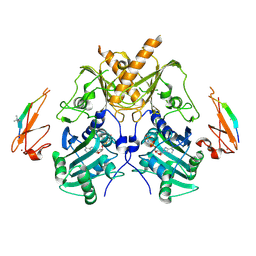 | | Crystal structure of mouse Morc3 ATPase-CW cassette in complex with AMPPNP and H3K4me3 peptide | | Descriptor: | MAGNESIUM ION, MORC family CW-type zinc finger protein 3, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Li, S, Du, J, Patel, D.J. | | Deposit date: | 2016-03-23 | | Release date: | 2016-08-17 | | Last modified: | 2016-09-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Mouse MORC3 is a GHKL ATPase that localizes to H3K4me3 marked chromatin
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
3QZS
 
 | |
