6IAC
 
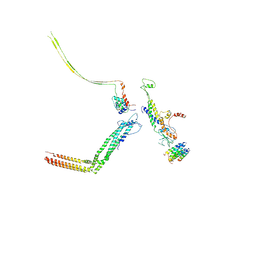 | | Portal and tail of native bacteriophage P68 | | Descriptor: | Lower collar protein, Minor structural protein, Portal protein, ... | | Authors: | Hrebik, D, Skubnik, K, Fuzik, T, Plevka, P. | | Deposit date: | 2018-11-26 | | Release date: | 2019-11-06 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure and genome ejection mechanism ofStaphylococcus aureusphage P68.
Sci Adv, 5, 2019
|
|
6IAW
 
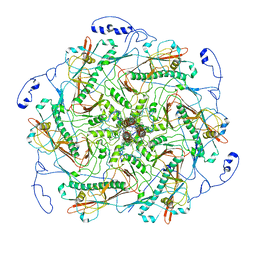 | | Structure of head fiber and inner core protein gp22 of native bacteriophage P68 | | Descriptor: | Arstotzka protein, Head fiber, Major head protein, ... | | Authors: | Hrebik, D, Skubnik, K, Fuzik, T, Plevka, P. | | Deposit date: | 2018-11-27 | | Release date: | 2019-11-06 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure and genome ejection mechanism ofStaphylococcus aureusphage P68.
Sci Adv, 5, 2019
|
|
1GSG
 
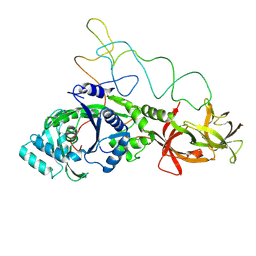 | | Structure of E.coli glutaminyl-tRNA synthetase complexed with trnagln and ATP at 2.8 Angstroms resolution | | Descriptor: | GLUTAMINYL-TRNA SYNTHETASE, TRNAGLN | | Authors: | Rould, M.A, Perona, J.J, Soell, D, Steitz, T.A. | | Deposit date: | 1990-04-03 | | Release date: | 1992-02-24 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of E. coli glutaminyl-tRNA synthetase complexed with tRNA(Gln) and ATP at 2.8 A resolution.
Science, 246, 1989
|
|
1AAW
 
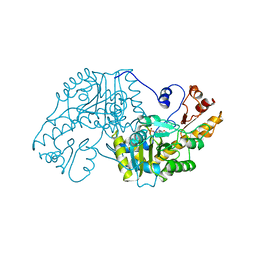 | | THE STRUCTURAL BASIS FOR THE ALTERED SUBSTRATE SPECIFICITY OF THE R292D ACTIVE SITE MUTANT OF ASPARTATE AMINOTRANSFERASE FROM E. COLI | | Descriptor: | ASPARTATE AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Almo, S.C, Smith, D.L, Danishefsky, A.T, Ringe, D. | | Deposit date: | 1993-07-13 | | Release date: | 1993-10-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structural basis for the altered substrate specificity of the R292D active site mutant of aspartate aminotransferase from E. coli.
Protein Eng., 7, 1994
|
|
9IN9
 
 | | Structure of the viral channelrhodopsin OLPVR1 at low pH obtained by soaking | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-hexadec-9-enoate, EICOSANE, ... | | Authors: | Bukhdruker, S, Zabelskii, D, Astashkin, R, Gordeliy, V. | | Deposit date: | 2024-07-05 | | Release date: | 2025-03-12 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Ion-conducting and gating molecular mechanisms of channelrhodopsin revealed by true-atomic-resolution structures of open and closed states.
Nat.Struct.Mol.Biol., 2025
|
|
3RUB
 
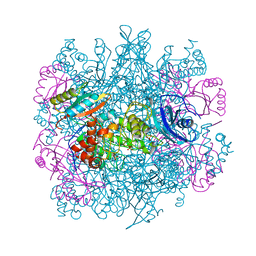 | | CRYSTAL STRUCTURE OF THE UNACTIVATED FORM OF RIBULOSE-1,5-BISPHOSPHATE CARBOXYLASE(SLASH)OXYGENASE FROM TOBACCO REFINED AT 2.0-ANGSTROMS RESOLUTION | | Descriptor: | ASPARAGINE, RIBULOSE 1,5-BISPHOSPHATE CARBOXYLASE/OXYGENASE, FORM III, ... | | Authors: | Schreuder, H, Cascio, D, Curmi, P.M.G, Chapman, M.S, Suh, S.W, Eisenberg, D.S. | | Deposit date: | 1990-05-25 | | Release date: | 1992-10-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the unactivated form of ribulose-1,5-bisphosphate carboxylase/oxygenase from tobacco refined at 2.0-A resolution.
J.Biol.Chem., 267, 1992
|
|
9IN7
 
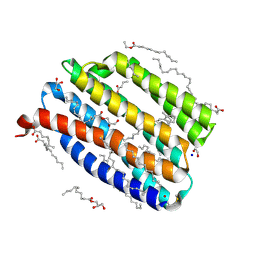 | | True-atomic resolution crystal structure of the closed state of the viral channelrhodopsin OLPVR1 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-hexadec-9-enoate, EICOSANE, ... | | Authors: | Bukhdruker, S, Zabelskii, D, Astashkin, R, Gordeliy, V. | | Deposit date: | 2024-07-05 | | Release date: | 2025-03-12 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Ion-conducting and gating molecular mechanisms of channelrhodopsin revealed by true-atomic-resolution structures of open and closed states.
Nat.Struct.Mol.Biol., 2025
|
|
9IN8
 
 | | True-atomic resolution crystal structure of the open state of the viral channelrhodopsin OLPVR1 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-hexadec-9-enoate, EICOSANE, ... | | Authors: | Bukhdruker, S, Zabelskii, D, Astashkin, R, Bourenkov, G, Gordeliy, V. | | Deposit date: | 2024-07-05 | | Release date: | 2025-03-12 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Ion-conducting and gating molecular mechanisms of channelrhodopsin revealed by true-atomic-resolution structures of open and closed states.
Nat.Struct.Mol.Biol., 2025
|
|
1H3O
 
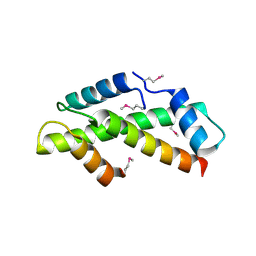 | | Crystal Structure of the Human TAF4-TAF12 (TAFII135-TAFII20) Complex | | Descriptor: | TRANSCRIPTION INITIATION FACTOR TFIID 135 KDA SUBUNIT, TRANSCRIPTION INITIATION FACTOR TFIID 20/15 KDA SUBUNITS | | Authors: | Werten, S, Mitschler, A, Moras, D. | | Deposit date: | 2002-09-12 | | Release date: | 2002-09-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of a Subcomplex of Human Transcription Factor TFIID Formed by TATA Binding Protein-Associated Factors Htaf4 (Htaf(II)135) and Htaf12 (Htaf(II)20).
J.Biol.Chem., 277, 2002
|
|
7W5L
 
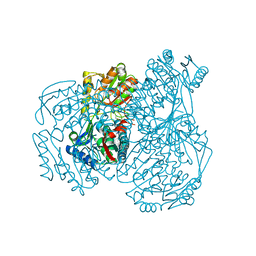 | | The crystal structure of the oxidized form of Gluconobacter oxydans WSH-004 SNDH | | Descriptor: | L-sorbosone dehydrogenase, NAD(P) dependent | | Authors: | Li, D, Hou, X.D, Rao, Y.J, Zhou, J.W, Chen, J. | | Deposit date: | 2021-11-30 | | Release date: | 2023-01-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Insight into the Catalytic Mechanisms of an L-Sorbosone Dehydrogenase.
Adv Sci, 10, 2023
|
|
1A2Y
 
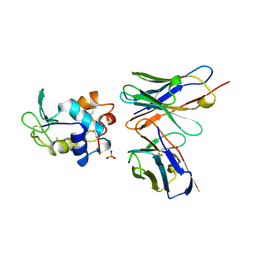 | | HEN EGG WHITE LYSOZYME, D18A MUTANT, IN COMPLEX WITH MOUSE MONOCLONAL ANTIBODY D1.3 | | Descriptor: | IGG1-KAPPA D1.3 FV (HEAVY CHAIN), IGG1-KAPPA D1.3 FV (LIGHT CHAIN), LYSOZYME, ... | | Authors: | Tsuchiya, D, Mariuzza, R.A. | | Deposit date: | 1998-01-13 | | Release date: | 1998-04-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A mutational analysis of binding interactions in an antigen-antibody protein-protein complex.
Biochemistry, 37, 1998
|
|
7UAF
 
 | | Human meprin alpha inhibitory complex with compound 10d (N~3~,N~3~-bis[(2H-1,3-benzodioxol-5-yl)methyl]-N-hydroxy-beta-alaninamide) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bayly-Jones, C, Lupton, C.J, Fritz, C, Schlenzig, D, Whisstock, J.C. | | Deposit date: | 2022-03-12 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Helical ultrastructure of the metalloprotease meprin alpha in complex with a small molecule inhibitor.
Nat Commun, 13, 2022
|
|
5L9U
 
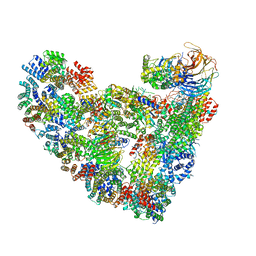 | | Model of human Anaphase-promoting complex/Cyclosome (APC/C-CDH1) with a cross linked Ubiquitin variant-substrate-UBE2C (UBCH10) complex representing key features of multiubiquitination | | Descriptor: | Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 10, Anaphase-promoting complex subunit 11, ... | | Authors: | Brown, N.G, VanderLinden, R, Dube, P, Haselbach, D, Peters, J.M, Stark, H, Schulman, B.A. | | Deposit date: | 2016-06-11 | | Release date: | 2016-09-14 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | Dual RING E3 Architectures Regulate Multiubiquitination and Ubiquitin Chain Elongation by APC/C.
Cell, 165, 2016
|
|
5ABV
 
 | |
7ZJF
 
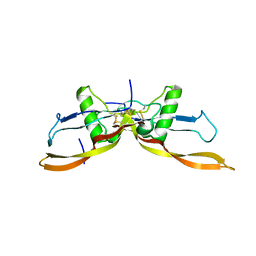 | | R399E, a mutated form of GDF5, for disease modification of osteoarthritis | | Descriptor: | Growth/differentiation factor 5 | | Authors: | Gigout, A, Wekmann, D, Menges, S, Brenneis, C, Henson, F, Cowan, K.J, Musil, D, Thudium, C, Guehring, H, Michaelis, M, Kleinschmidt-Doerr, K. | | Deposit date: | 2022-04-10 | | Release date: | 2022-09-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | R399E, A Mutated Form of Growth and Differentiation Factor 5, for Disease Modification of Osteoarthritis.
Arthritis Rheumatol, 75, 2023
|
|
8QW7
 
 | | Crystal Structure of compound 4 in complex with KRAS G12V C118S GDP and pVHL:ElonginC:ElonginB | | Descriptor: | (2S,4R)-1-[(2R)-2-[3-[4-[(3S)-4-[4-[5-[(4S)-2-azanyl-3-cyano-4-methyl-6,7-dihydro-5H-1-benzothiophen-4-yl]-1,2,4-oxadiazol-3-yl]pyrimidin-2-yl]-3-methyl-1,4-diazepan-1-yl]butoxy]-1,2-oxazol-5-yl]-3-methyl-butanoyl]-N-[[4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Elongin-B, Elongin-C, ... | | Authors: | Zollman, D, Farnaby, W, Ciulli, A. | | Deposit date: | 2023-10-18 | | Release date: | 2023-12-06 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Targeting cancer with small-molecule pan-KRAS degraders.
Science, 385, 2024
|
|
8QVU
 
 | | Crystal Structure of ligand ACBI3 in complex with KRAS G12D C118S GDP and pVHL:ElonginC:ElonginB complex | | Descriptor: | (2S,4R)-1-[(2S)-2-[4-[4-[(3S)-4-[4-[5-[(4S)-2-azanyl-3-cyano-4-methyl-6,7-dihydro-5H-1-benzothiophen-4-yl]-1,2,4-oxadiazol-3-yl]pyrimidin-2-yl]-3-methyl-1,4-diazepan-1-yl]butoxy]-1,2,3-triazol-1-yl]-3-methyl-butanoyl]-N-[(1R)-1-[4-(4-methyl-1,3-thiazol-5-yl)phenyl]-2-oxidanyl-ethyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Elongin-B, Elongin-C, ... | | Authors: | Wijaya, A.J, Zollman, D, Farnaby, W, Ciulli, A. | | Deposit date: | 2023-10-18 | | Release date: | 2023-12-06 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Targeting cancer with small-molecule pan-KRAS degraders.
Science, 385, 2024
|
|
6JHK
 
 | |
8QU8
 
 | | PROTAC-mediated complex of KRAS with VHL/Elongin-B/Elongin-C/Cullin-2/Rbx1 | | Descriptor: | (2S,4R)-1-[(2S)-2-[4-[4-[(3S)-4-[4-[5-[(4S)-2-azanyl-3-cyano-4-methyl-6,7-dihydro-5H-1-benzothiophen-4-yl]-1,2,4-oxadiazol-3-yl]pyrimidin-2-yl]-3-methyl-1,4-diazepan-1-yl]butoxy]-1,2,3-triazol-1-yl]-3-methyl-butanoyl]-N-[(1R)-1-[4-(4-methyl-1,3-thiazol-5-yl)phenyl]-2-oxidanyl-ethyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Cullin-2, E3 ubiquitin-protein ligase RBX1, ... | | Authors: | Fischer, G, Peter, D, Arce-Solano, S. | | Deposit date: | 2023-10-14 | | Release date: | 2023-12-06 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Targeting cancer with small-molecule pan-KRAS degraders.
Science, 385, 2024
|
|
8QW6
 
 | | Crystal Structure of compound 3 in complex with KRAS G12V C118S GDP and pVHL:ElonginC:ElonginB | | Descriptor: | (2S,4R)-1-[(2S)-2-[6-[(3S)-4-[4-[5-[(4S)-2-azanyl-3-cyano-4-methyl-6,7-dihydro-5H-1-benzothiophen-4-yl]-1,2,4-oxadiazol-3-yl]pyrimidin-2-yl]-3-methyl-1,4-diazepan-1-yl]hexanoylamino]-3,3-dimethyl-butanoyl]-N-[[4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Elongin-B, Elongin-C, ... | | Authors: | Zollman, D, Farnaby, W, Ciulli, A. | | Deposit date: | 2023-10-18 | | Release date: | 2023-12-06 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Targeting cancer with small-molecule pan-KRAS degraders.
Science, 385, 2024
|
|
1H6B
 
 | |
1GZN
 
 | | Structure of PKB kinase domain | | Descriptor: | RAC-BETA SERINE/THREONINE PROTEIN KINASE | | Authors: | Barford, D, Yang, J, Hemmings, B.A. | | Deposit date: | 2002-05-24 | | Release date: | 2003-05-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular Mechanism for the Regulation of Protein Kinase B/Akt by Hydrophobic Motif Phosphorylation
Mol.Cell, 9, 2002
|
|
1H6C
 
 | |
1CF4
 
 | | CDC42/ACK GTPASE-BINDING DOMAIN COMPLEX | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, PROTEIN (ACTIVATED P21CDC42HS KINASE), ... | | Authors: | Mott, H.R, Owen, D, Nietlispach, D, Lowe, P.N, Lim, L, Laue, E.D. | | Deposit date: | 1999-03-23 | | Release date: | 1999-06-18 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure of the small G protein Cdc42 bound to the GTPase-binding domain of ACK.
Nature, 399, 1999
|
|
1H4M
 
 | | Sulfurtransferase from Azotobacter vinelandii in complex with phosphate | | Descriptor: | 1,2-ETHANEDIOL, PUTATIVE THIOSULFATE SULFURTRANSFERASE | | Authors: | Bordo, D, Forlani, F, Spallarossa, A, Colnaghi, R, Carpen, A, Pagani, S, Bolognesi, M. | | Deposit date: | 2001-05-11 | | Release date: | 2002-05-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Persulfurated Cysteine Promotes Active Site Reactivity in Azotobacter Vinelandii Rhodanse
Biol.Chem., 382, 2001
|
|
