7C6U
 
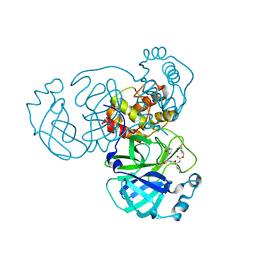 | | Crystal structure of SARS-CoV-2 complexed with GC376 | | Descriptor: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase | | Authors: | Fu, L, Feng, Y, Qi, J, Gao, F.G. | | Deposit date: | 2020-05-22 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Both Boceprevir and GC376 efficaciously inhibit SARS-CoV-2 by targeting its main protease.
Nat Commun, 11, 2020
|
|
7C4T
 
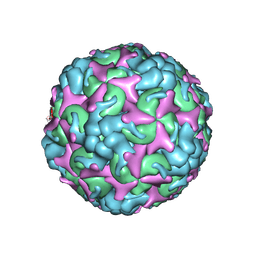 | | Cryo-EM structure of A particle Coxsackievirus A10 at pH 7.4 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3 | | Authors: | Cui, Y, Peng, R, Song, H, Tong, Z, Gao, G.F, Qi, J. | | Deposit date: | 2020-05-18 | | Release date: | 2020-07-22 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Molecular basis of Coxsackievirus A10 entry using the two-in-one attachment and uncoating receptor KRM1.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7C4W
 
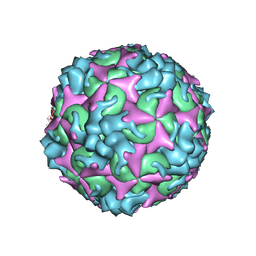 | | Cryo-EM structure of A particle Coxsackievirus A10 at pH 5.5 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3 | | Authors: | Cui, Y, Peng, R, Song, H, Tong, Z, Gao, G.F, Qi, J. | | Deposit date: | 2020-05-18 | | Release date: | 2020-07-22 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular basis of Coxsackievirus A10 entry using the two-in-one attachment and uncoating receptor KRM1.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7E9W
 
 | | The Crystal Structure of D-psicose-3-epimerase from Biortus. | | Descriptor: | D-psicose 3-epimerase, GLYCEROL, MANGANESE (II) ION | | Authors: | Wang, F, Xu, C, Qi, J, Zhang, M, Tian, F, Wang, M. | | Deposit date: | 2021-03-05 | | Release date: | 2021-03-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Crystal Structure of D-psicose-3-epimerase from Biortus.
To Be Published
|
|
7E57
 
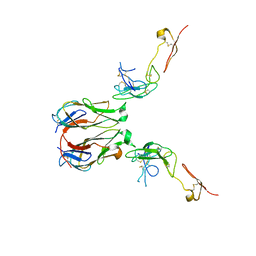 | | Crystal structure of murine GITR-GITRL complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Tumor necrosis factor ligand superfamily member 18, Tumor necrosis factor receptor superfamily member 18, ... | | Authors: | Zhao, M, Tan, S, Fu, L, Chai, Y, Qi, J, Gao, G.F. | | Deposit date: | 2021-02-18 | | Release date: | 2021-09-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.302 Å) | | Cite: | Atypical TNF-TNFR superfamily binding interface in the GITR-GITRL complex for T cell activation.
Cell Rep, 36, 2021
|
|
7E5M
 
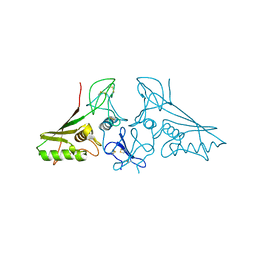 | | crystal structure of trans assembled human TROP-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Tumor-associated calcium signal transducer 2 | | Authors: | Sun, M, Zhang, H, Chai, Y, Qi, J, Gao, G.F, Tan, S. | | Deposit date: | 2021-02-19 | | Release date: | 2021-12-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insights into the cis and trans assembly of human trophoblast cell surface antigen 2.
Iscience, 24, 2021
|
|
7E5N
 
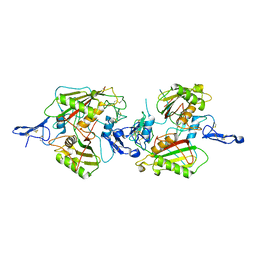 | | crystal structure of cis assembled TROP-2 | | Descriptor: | Tumor-associated calcium signal transducer 2 | | Authors: | Sun, M, Zhang, H, Chai, Y, Qi, J, Gao, G.F, Tan, S. | | Deposit date: | 2021-02-19 | | Release date: | 2021-12-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insights into the cis and trans assembly of human trophoblast cell surface antigen 2.
Iscience, 24, 2021
|
|
7ESE
 
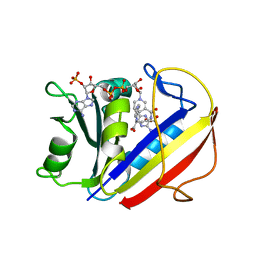 | | The Crystal Structure of human DHFR from Biortus | | Descriptor: | Dihydrofolate reductase, FOLIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Wang, F, Cheng, W, Xu, C, Qi, J, Bao, X, Miao, Q. | | Deposit date: | 2021-05-10 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Crystal Structure of human DHFR from Biortus
To Be Published
|
|
7F9L
 
 | | Crystal structure of the variable region of Plasmodium RIFIN #6 (PF3D7_1400600) in complex with LAIR1 (with T67L, N69S and A77T mutations) | | Descriptor: | Leukocyte-associated immunoglobulin-like receptor 1, Rifin | | Authors: | Xie, Y, Song, H, Li, X, Qi, J, Gao, G.F. | | Deposit date: | 2021-07-04 | | Release date: | 2021-08-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of malarial parasite RIFIN-mediated immune escape against LAIR1.
Cell Rep, 36, 2021
|
|
7F9N
 
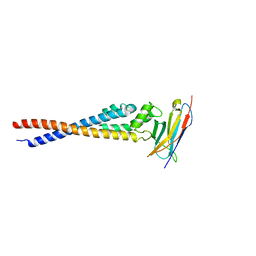 | | Crystal structure of the variable region of Plasmodium RIFIN #4 (PF3D7_1000500) in complex with LAIR1 | | Descriptor: | Leukocyte-associated immunoglobulin-like receptor 1, Rifin | | Authors: | Xie, Y, Song, H, Li, X, Qi, J, Gao, G.F. | | Deposit date: | 2021-07-04 | | Release date: | 2021-08-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of malarial parasite RIFIN-mediated immune escape against LAIR1.
Cell Rep, 36, 2021
|
|
7F9K
 
 | | Crystal structure of the variable region of Plasmodium RIFIN #6(PF3D7_1400600) | | Descriptor: | Rifin | | Authors: | Xie, Y, Song, H, Li, X, Qi, J, Gao, G.F. | | Deposit date: | 2021-07-04 | | Release date: | 2021-08-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structural basis of malarial parasite RIFIN-mediated immune escape against LAIR1.
Cell Rep, 36, 2021
|
|
7F9M
 
 | | Crystal structure of the variable region of Plasmodium RIFIN #4 (PF3D7_1000500) in complex with LAIR1 (with T67L, N69S and A77T mutations) | | Descriptor: | Leukocyte-associated immunoglobulin-like receptor 1, Rifin | | Authors: | Xie, Y, Song, H, Li, X, Qi, J, Gao, G.F. | | Deposit date: | 2021-07-04 | | Release date: | 2021-08-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of malarial parasite RIFIN-mediated immune escape against LAIR1.
Cell Rep, 36, 2021
|
|
6IEX
 
 | | Crystal structure of HLA-B*4001 in complex with SARS-CoV derived peptide N216-225 GETALALLLL | | Descriptor: | Beta-2-microglobulin, GLY-GLU-THR-ALA-LEU-ALA-LEU-LEU-LEU-LEU, MHC class I antigen | | Authors: | Ji, W, Niu, L, Peng, W, Zhang, Y, Shi, Y, Qi, J, Gao, G.F, Liu, W.J. | | Deposit date: | 2018-09-17 | | Release date: | 2019-09-18 | | Last modified: | 2021-03-31 | | Method: | X-RAY DIFFRACTION (2.314 Å) | | Cite: | Salt bridge-forming residues positioned over viral peptides presented by MHC class I impacts T-cell recognition in a binding-dependent manner.
Mol.Immunol., 112, 2019
|
|
6II9
 
 | | Crystal structure of H7 hemagglutinin from A/Anhui/1/2013 in complex with a human neutralizing antibody L3A-44 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of L3A-44 Fab, Hemagglutinin, ... | | Authors: | Jiang, H.H, Shi, Y, Qi, J, Gao, G.F. | | Deposit date: | 2018-10-03 | | Release date: | 2018-10-24 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure-function analysis of neutralizing antibodies to H7N9 influenza from naturally infected humans.
Nat Microbiol, 4, 2019
|
|
6II8
 
 | | Crystal structure of H7 hemagglutinin from A/Anhui/1/2013 in complex with a human neutralizing antibody L4B-18 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of L4B-18 Fab, Hemagglutinin, ... | | Authors: | Jiang, H.H, Shi, Y, Qi, J, Gao, G.F. | | Deposit date: | 2018-10-03 | | Release date: | 2018-10-24 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.32 Å) | | Cite: | Structure-function analysis of neutralizing antibodies to H7N9 influenza from naturally infected humans.
Nat Microbiol, 4, 2019
|
|
6II4
 
 | | Crystal structure of H7 hemagglutinin from A/Anhui/1/2013 in complex with a human neutralizing antibody L4A-14 | | Descriptor: | Heavy chain of L4A-14 Fab, Hemagglutinin, Light chain of L4A-14 Fab | | Authors: | Jiang, H.H, Shi, Y, Qi, J, Gao, G.F. | | Deposit date: | 2018-10-03 | | Release date: | 2018-10-24 | | Last modified: | 2019-02-06 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure-function analysis of neutralizing antibodies to H7N9 influenza from naturally infected humans.
Nat Microbiol, 4, 2019
|
|
6ISC
 
 | | complex structure of mCD226-ecto and hCD155-D1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CD226 antigen, Poliovirus receptor | | Authors: | Wang, H, Qi, J, Zhang, S, Li, Y, Tan, S, Gao, G.F. | | Deposit date: | 2018-11-16 | | Release date: | 2018-12-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Binding mode of the side-by-side two-IgV molecule CD226/DNAM-1 to its ligand CD155/Necl-5.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6ISA
 
 | | mCD226 | | Descriptor: | CD226 antigen | | Authors: | Wang, H, Qi, J, Zhang, S, Li, Y, Tan, S, Gao, G.F. | | Deposit date: | 2018-11-16 | | Release date: | 2018-12-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Binding mode of the side-by-side two-IgV molecule CD226/DNAM-1 to its ligand CD155/Necl-5.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6ISB
 
 | | crystal structure of human CD226 | | Descriptor: | CD226 antigen | | Authors: | Wang, H, Qi, J, Zhang, S, Li, Y, Tan, S, Gao, G.F. | | Deposit date: | 2018-11-16 | | Release date: | 2018-12-26 | | Last modified: | 2019-02-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Binding mode of the side-by-side two-IgV molecule CD226/DNAM-1 to its ligand CD155/Necl-5.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6KY8
 
 | | Crystal structure of ASFV dUTPase | | Descriptor: | E165R | | Authors: | Li, C, Chai, Y, Song, H, Qi, J, Sun, Y, Gao, G.F. | | Deposit date: | 2019-09-17 | | Release date: | 2019-11-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Crystal Structure of African Swine Fever Virus dUTPase Reveals a Potential Drug Target.
Mbio, 10, 2019
|
|
6KY9
 
 | | Crystal structure of ASFV dUTPase and UMP complex | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, E165R | | Authors: | Li, C, Chai, Y, Song, H, Qi, J, Sun, Y, Gao, G.F. | | Deposit date: | 2019-09-17 | | Release date: | 2019-11-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of African Swine Fever Virus dUTPase Reveals a Potential Drug Target.
Mbio, 10, 2019
|
|
6BA5
 
 | |
6BAN
 
 | |
3AL4
 
 | | Crystal structure of the swine-origin A (H1N1)-2009 influenza A virus hemagglutinin (HA) reveals similar antigenicity to that of the 1918 pandemic virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | Zhang, W, Qi, J.X, Shi, Y, Li, Q, Yan, J.H, Gao, G.F. | | Deposit date: | 2010-07-22 | | Release date: | 2010-08-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.872 Å) | | Cite: | Crystal structure of the swine-origin A (H1N1)-2009 influenza A virus hemagglutinin (HA) reveals similar antigenicity to that of the 1918 pandemic virus
Protein Cell, 1, 2010
|
|
6BGI
 
 | | Cryo-EM structure of the TMEM16A calcium-activated chloride channel in nanodisc | | Descriptor: | Anoctamin-1, CALCIUM ION | | Authors: | Dang, S, Feng, S, Tien, J, Peters, C.J, Bulkley, D, Lolicato, M, Zhao, J, Zuberbuhler, K, Ye, W, Qi, J, Chen, T, Craik, C.S, Jan, Y.N, Minor Jr, D.L, Cheng, Y, Jan, L.Y. | | Deposit date: | 2017-10-28 | | Release date: | 2017-12-27 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures of the TMEM16A calcium-activated chloride channel.
Nature, 552, 2017
|
|
