1UE2
 
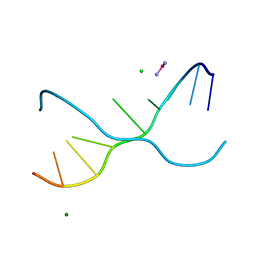 | | Crystal structure of d(GC38GAAAGCT) | | Descriptor: | 5'-D(*GP*(C38)P*GP*AP*AP*AP*GP*CP*T)-3', CHLORIDE ION, COBALT HEXAMMINE(III), ... | | Authors: | Sunami, T, Kondo, J, Hirao, I, Watanaba, K, Miura, K, Takenaka, A. | | Deposit date: | 2003-05-08 | | Release date: | 2004-01-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of d(GCGAAAGC) (hexagonal form): a base-intercalated duplex as a stable structure.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
6JT4
 
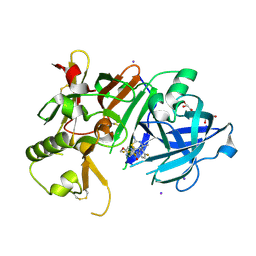 | | Crystal Structure of BACE1 in complex with N-{3-[(4S,6S)-2-amino-4-methyl-6-(trifluoromethyl)-5,6-dihydro-4H-1,3-thiazin-4-yl]-4-fluorophenyl}-5-(fluoromethoxy)pyrazine-2-carboxamide | | Descriptor: | Beta-secretase 1, GLYCEROL, IODIDE ION, ... | | Authors: | Anan, K, Iso, Y, Oguma, T, Nakahara, K, Suzuki, S, Yamamoto, T, Matsuoka, E, Ito, H, Sakaguchi, G, Ando, S, Morimoto, K, Kanegawa, N, Kido, Y, Kawachi, T, Fukushima, T, Teisman, A, Urmaliya, V, Dhuyvetter, D, Borghys, H, Austin, N, Bergh, A.V.D, Verboven, P, Bischoff, F, Gijsen, H.J.M, Yamano, Y, Kusakabe, K.I. | | Deposit date: | 2019-04-08 | | Release date: | 2020-01-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Trifluoromethyl Dihydrothiazine-Based beta-Secretase (BACE1) Inhibitors with Robust Central beta-Amyloid Reduction and Minimal Covalent Binding Burden.
Chemmedchem, 14, 2019
|
|
1D6X
 
 | |
7INS
 
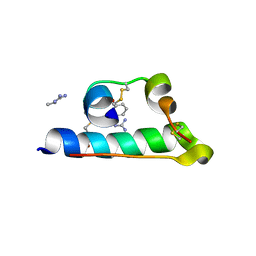 | | STRUCTURE OF PORCINE INSULIN COCRYSTALLIZED WITH CLUPEINE Z | | Descriptor: | GENERAL PROTAMINE CHAIN, INSULIN (CHAIN A), INSULIN (CHAIN B), ... | | Authors: | Balschmidt, P, Hansen, F.B, Dodson, E, Dodson, G, Korber, F. | | Deposit date: | 1991-09-03 | | Release date: | 1994-01-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of porcine insulin cocrystallized with clupeine Z.
Acta Crystallogr.,Sect.B, 47, 1991
|
|
5YGY
 
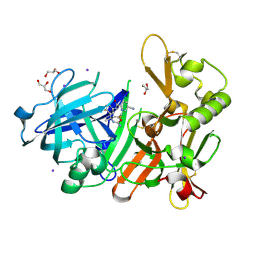 | | Crystal Structure of BACE1 in complex with (S)-N-(3-(2-amino-6-(fluoromethyl)-4 -methyl-4H-1,3-oxazin-4-yl)-4-fluorophenyl)-5-cyanopicolinamide | | Descriptor: | Beta-secretase 1, GLYCEROL, IODIDE ION, ... | | Authors: | Fuchino, K, Mitsuoka, Y, Masui, M, Kurose, N, Yoshida, S, Komano, K, Yamamoto, T, Ogawa, M, Unemura, C, Hosono, M, Ito, H, Sakaguchi, G, Ando, S, Ohnishi, S, Kido, Y, Fukushima, T, Miyajima, H, Hiroyama, S, Koyabu, K, Dhuyvetter, D, Borghys, H, Gijsen, H, Yamano, Y, Iso, Y, Kusakabe, K. | | Deposit date: | 2017-09-27 | | Release date: | 2018-05-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Rational Design of Novel 1,3-Oxazine Based beta-Secretase (BACE1) Inhibitors: Incorporation of a Double Bond To Reduce P-gp Efflux Leading to Robust A beta Reduction in the Brain
J. Med. Chem., 61, 2018
|
|
4Z16
 
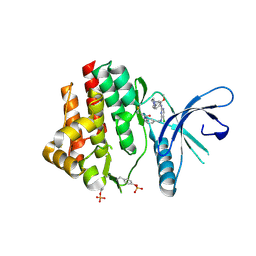 | | Crystal Structure of the Jak3 Kinase Domain Covalently Bound to N-(3-(((5-chloro-2-((2-methoxy-4-(4-methylpiperazin-1-yl)phenyl)amino)pyrimidin-4-yl)amino)methyl)phenyl)acrylamide | | Descriptor: | N-(3-{[(5-chloro-2-{[2-methoxy-4-(4-methylpiperazin-1-yl)phenyl]amino}pyrimidin-4-yl)amino]methyl}phenyl)prop-2-enamide, Tyrosine-protein kinase JAK3 | | Authors: | McNally, R, Tan, L, Gray, N.S, Eck, M.J. | | Deposit date: | 2015-03-26 | | Release date: | 2016-02-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Development of Selective Covalent Janus Kinase 3 Inhibitors.
J.Med.Chem., 58, 2015
|
|
5YGX
 
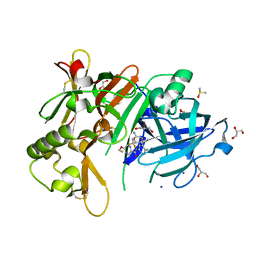 | | Structure of BACE1 in complex with N-(3-((4R,5R,6S)-2-amino-6-(1,1-difluoroethyl)-5-fluoro-4-methyl-5,6-dihydro-4H-1,3-oxazin-4-yl)-4-fluorophenyl)-5-(fluoromethoxy)pyrazine-2-carboxamide | | Descriptor: | Beta-secretase 1, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Nakahara, K, Fuchino, K, Komano, K, Asada, N, Tadano, G, Hasegawa, T, Yamamoto, T, Sako, Y, Ogawa, M, Unemura, C, Hosono, M, Sakaguchi, G, Ando, S, Ohnishi, S, Kido, Y, Fukushima, T, Dhuyvetter, D, Borghys, H, Gijsen, H, Yamano, Y, Iso, Y, Kusakabe, K. | | Deposit date: | 2017-09-27 | | Release date: | 2018-08-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of Potent and Centrally Active 6-Substituted 5-Fluoro-1,3-dihydro-oxazine beta-Secretase (BACE1) Inhibitors via Active Conformation Stabilization
J. Med. Chem., 61, 2018
|
|
1ZNI
 
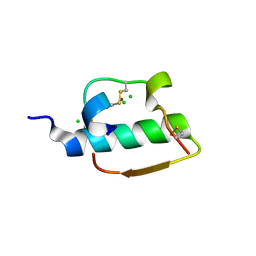 | | INSULIN | | Descriptor: | CHLORIDE ION, INSULIN, ZINC ION | | Authors: | Turkenburg, M.G.W, Whittingham, J.L, Dodson, G.G, Dodson, E.J, Xiao, B, Bentley, G.A. | | Deposit date: | 1997-09-23 | | Release date: | 1998-01-28 | | Last modified: | 2018-07-04 | | Method: | X-RAY DIFFRACTION (1.498 Å) | | Cite: | Structure of insulin in 4-zinc insulin.
Nature, 261, 1976
|
|
1IZB
 
 | |
1IZA
 
 | |
1BEN
 
 | | INSULIN COMPLEXED WITH 4-HYDROXYBENZAMIDE | | Descriptor: | 4-HYDROXYBENZAMIDE, CHLORIDE ION, HUMAN INSULIN, ... | | Authors: | Smith, G.D, Ciszak, E, Pangborn, W. | | Deposit date: | 1996-02-15 | | Release date: | 1996-07-11 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A novel complex of a phenolic derivative with insulin: structural features related to the T-->R transition.
Protein Sci., 5, 1996
|
|
5ZSU
 
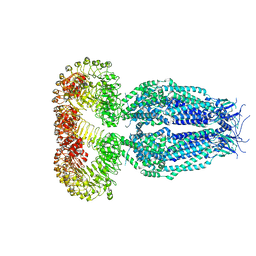 | | Structure of the human homo-hexameric LRRC8A channel at 4.25 Angstroms | | Descriptor: | Volume-regulated anion channel subunit LRRC8A | | Authors: | Kasuya, G, Nakane, T, Yokoyama, T, Shirouzu, M, Ishitani, R, Nureki, O. | | Deposit date: | 2018-04-29 | | Release date: | 2018-08-15 | | Last modified: | 2018-09-26 | | Method: | ELECTRON MICROSCOPY (4.25 Å) | | Cite: | Cryo-EM structures of the human volume-regulated anion channel LRRC8.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6FHV
 
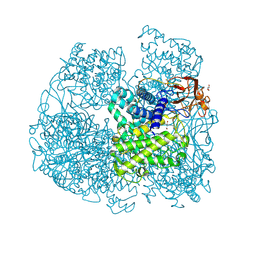 | | Crystal structure of Penicillium oxalicum Glucoamylase | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Roth, C, Moroz, O.V, Ariza, A, Friis, E.P, Davies, G.J, Wilson, K.S. | | Deposit date: | 2018-01-15 | | Release date: | 2018-05-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insight into industrially relevant glucoamylases: flexible positions of starch-binding domains.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6FNY
 
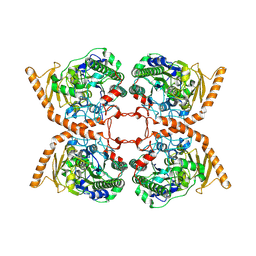 | | CRYSTAL STRUCTURE OF A CHOLINE SULFATASE FROM SINORHIZOBIUM MELLILOTI | | Descriptor: | CALCIUM ION, Choline-sulfatase | | Authors: | Valkov, E, Van Loo, B, Hollfelder, F, Hyvonen, M. | | Deposit date: | 2018-02-05 | | Release date: | 2018-02-28 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural and Mechanistic Analysis of the Choline Sulfatase from Sinorhizobium melliloti: A Class I Sulfatase Specific for an Alkyl Sulfate Ester.
J. Mol. Biol., 430, 2018
|
|
6FTN
 
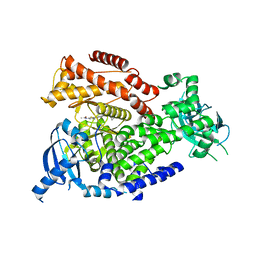 | | mPI3Kd IN COMPLEX WITH AZ2 | | Descriptor: | Phosphor inositol 3 kinase, ~{N}-[5-[2-[(1~{S})-1-cyclopropylethyl]-7-methyl-1-oxidanylidene-3~{H}-isoindol-5-yl]-4-methyl-1,3-thiazol-2-yl]ethanamide | | Authors: | Petersen, J. | | Deposit date: | 2018-02-22 | | Release date: | 2018-06-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Highly Isoform Selective Orally Bioavailable Phosphoinositide 3-Kinase (PI3K)-gamma Inhibitors.
J. Med. Chem., 61, 2018
|
|
6FRV
 
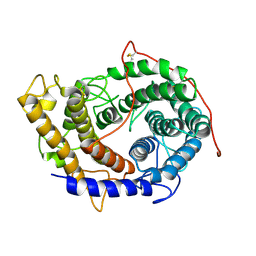 | | Structure of the catalytic domain of Aspergillus niger Glucoamylase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glucoamylase, ... | | Authors: | Roth, C, Moroz, O.V, Ariza, A, Friis, E.P, Davies, G.J, Wilson, K.S. | | Deposit date: | 2018-02-16 | | Release date: | 2018-05-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insight into industrially relevant glucoamylases: flexible positions of starch-binding domains.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6GY0
 
 | | mPI3Kd IN COMPLEX WITH AZ3 | | Descriptor: | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform, ~{N}-[4-methyl-5-(1-oxidanylidene-7-sulfamoyl-isoindol-5-yl)-1,3-thiazol-2-yl]ethanamide | | Authors: | Petersen, J. | | Deposit date: | 2018-06-27 | | Release date: | 2019-02-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | A class of highly selective inhibitors bind to an active state of PI3K gamma.
Nat.Chem.Biol., 15, 2019
|
|
6FHW
 
 | | Structure of Hormoconis resinae Glucoamylase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Roth, C, Moroz, O.V, Ariza, A, Friis, E.P, Davies, G.J, Wilson, K.S. | | Deposit date: | 2018-01-15 | | Release date: | 2018-05-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural insight into industrially relevant glucoamylases: flexible positions of starch-binding domains.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
7EF8
 
 | | Crystal structure of mouse MUTYH in complex with DNA containing AP site analogue:8-oxoG (Form I) | | Descriptor: | Adenine DNA glycosylase, DNA (5'-D(*TP*AP*GP*TP*CP*CP*CP*(3DR)P*GP*TP*CP*TP*C)-3'), DNA (5'-D(*TP*GP*AP*GP*AP*CP*(8OG)P*GP*GP*GP*AP*CP*T)-3'), ... | | Authors: | Nakamura, T, Nakabeppu, Y, Yamagata, Y. | | Deposit date: | 2021-03-21 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of the mammalian adenine DNA glycosylase MUTYH: insights into the base excision repair pathway and cancer.
Nucleic Acids Res., 49, 2021
|
|
7EFA
 
 | |
7EF9
 
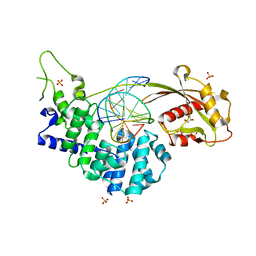 | | Crystal structure of mouse MUTYH in complex with DNA containing AP site analogue:8-oxoG (Form II) | | Descriptor: | Adenine DNA glycosylase, DNA (5'-D(*AP*TP*GP*AP*GP*AP*CP*(8OG)P*GP*GP*GP*AP*CP*T)-3'), DNA (5'-D(*TP*AP*GP*TP*CP*CP*CP*(3DR)P*GP*TP*CP*TP*C)-3'), ... | | Authors: | Nakamura, T, Nakabeppu, Y, Yamagata, Y. | | Deposit date: | 2021-03-21 | | Release date: | 2021-06-23 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structure of the mammalian adenine DNA glycosylase MUTYH: insights into the base excision repair pathway and cancer.
Nucleic Acids Res., 49, 2021
|
|
5GHA
 
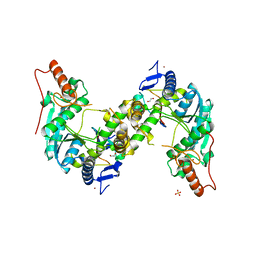 | | Sulfur Transferase TtuA in complex with Sulfur Carrier TtuB | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Sulfur Carrier TtuB, ... | | Authors: | Chen, M, Narai, S, Tanaka, Y, Yao, M. | | Deposit date: | 2016-06-19 | | Release date: | 2017-05-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Biochemical and structural characterization of oxygen-sensitive 2-thiouridine synthesis catalyzed by an iron-sulfur protein TtuA
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
3RGA
 
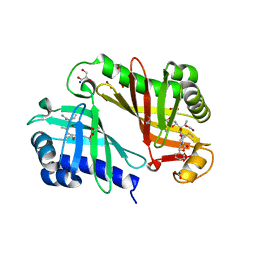 | | Crystal structure of epoxide hydrolase for polyether lasalocid A biosynthesis | | Descriptor: | (4R,5S)-3-[(2R)-2-{(2S,2'R,4S,5S,5'R)-2,5'-diethyl-5'-[(1S)-1-hydroxyethyl]-4-methyloctahydro-2,2'-bifuran-5-yl}butanoyl]-4-methyl-5-phenyl-1,3-oxazolidin-2-one, (4R,5S)-3-[(2R,3S,4S)-2-ethyl-5-[(3R)-2-ethyl-3-[2-[(2R,3R)-2-ethyl-3-methyl-oxiran-2-yl]ethyl]oxiran-2-yl]-3-hydroxy-4-methyl-pentanoyl]-4-methyl-5-phenyl-1,3-oxazolidin-2-one, ACETATE ION, ... | | Authors: | Hotta, K, Mathews, I.I, Chen, X, Kim, C.-Y. | | Deposit date: | 2011-04-08 | | Release date: | 2012-03-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Enzymatic catalysis of anti-Baldwin ring closure in polyether biosynthesis
Nature, 483, 2012
|
|
5B4E
 
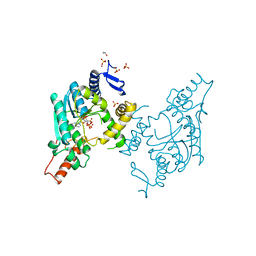 | | Sulfur Transferase TtuA in complex with iron sulfur cluster and ATP derivative | | Descriptor: | IRON/SULFUR CLUSTER, ISOPROPYL ALCOHOL, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Chen, M, Narai, S, Tanaka, Y, Yao, M. | | Deposit date: | 2016-04-03 | | Release date: | 2017-05-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.698 Å) | | Cite: | Biochemical and structural characterization of oxygen-sensitive 2-thiouridine synthesis catalyzed by an iron-sulfur protein TtuA
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5B4F
 
 | | Sulfur Transferase TtuA in complex with iron sulfur cluster | | Descriptor: | IRON/SULFUR CLUSTER, SULFATE ION, Sulfur Transferase TtuA, ... | | Authors: | Chen, M, Narai, S, Tanaka, Y, Yao, M. | | Deposit date: | 2016-04-03 | | Release date: | 2017-05-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Biochemical and structural characterization of oxygen-sensitive 2-thiouridine synthesis catalyzed by an iron-sulfur protein TtuA
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
