3WY1
 
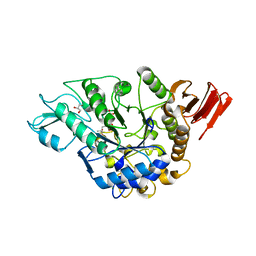 | | Crystal structure of alpha-glucosidase | | Descriptor: | (3R,5R,7R)-octane-1,3,5,7-tetracarboxylic acid, Alpha-glucosidase, GLYCEROL, ... | | Authors: | Shen, X, Gai, Z, Kato, K, Yao, M. | | Deposit date: | 2014-08-18 | | Release date: | 2015-06-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural analysis of the alpha-glucosidase HaG provides new insights into substrate specificity and catalytic mechanism
Acta Crystallogr. D Biol. Crystallogr., 71, 2015
|
|
2PV6
 
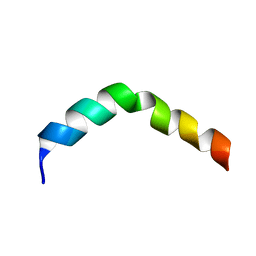 | | HIV-1 gp41 Membrane Proximal Ectodomain Region peptide in DPC micelle | | Descriptor: | Envelope glycoprotein | | Authors: | Sun, Z.-Y.J, Oh, K.J, Kim, M, Reinherz, E.L, Wagner, G. | | Deposit date: | 2007-05-09 | | Release date: | 2008-03-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | HIV-1 broadly neutralizing antibody extracts its epitope from a kinked gp41 ectodomain region on the viral membrane
Immunity, 28, 2008
|
|
3WGN
 
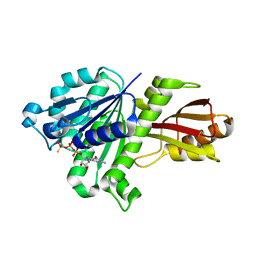 | | STAPHYLOCOCCUS AUREUS FTSZ bound with GTP-gamma-S | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Cell division protein FtsZ | | Authors: | Matsui, T, Mogi, N, Tanaka, I, Yao, M. | | Deposit date: | 2013-08-06 | | Release date: | 2013-12-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.606 Å) | | Cite: | Structural change in FtsZ Induced by intermolecular interactions between bound GTP and the T7 loop
J.Biol.Chem., 289, 2014
|
|
3WY3
 
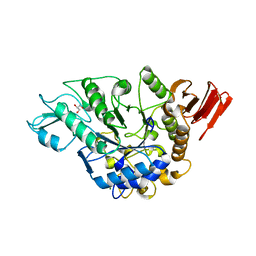 | | Crystal structure of alpha-glucosidase mutant D202N in complex with glucose and glycerol | | Descriptor: | Alpha-glucosidase, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Shen, X, Gai, Z, Kato, K, Yao, M. | | Deposit date: | 2014-08-18 | | Release date: | 2015-06-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural analysis of the alpha-glucosidase HaG provides new insights into substrate specificity and catalytic mechanism
Acta Crystallogr. D Biol. Crystallogr., 71, 2015
|
|
5XZI
 
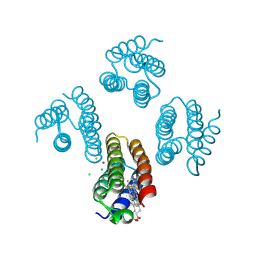 | |
7TTX
 
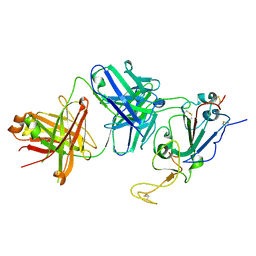 | |
7TTY
 
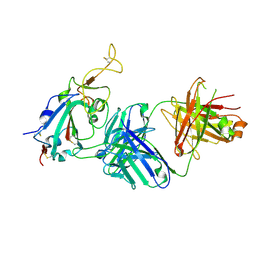 | |
5XZJ
 
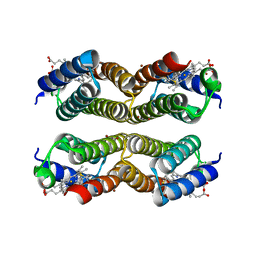 | |
8JT6
 
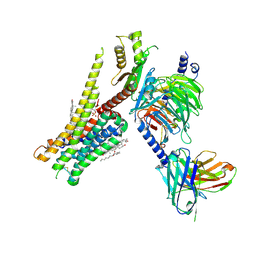 | | 5-HT1A-Gi in complex with compound (R)-IHCH-7179 | | Descriptor: | 1-(4-fluorophenyl)-4-[(7R)-2,5,11-triazatetracyclo[7.6.1.0^2,7.0^12,16]hexadeca-1(15),9,12(16),13-tetraen-5-yl]butan-1-one, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Chen, Z, Xu, P, Huang, S, Xu, H.E, Wang, S. | | Deposit date: | 2023-06-21 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Flexible scaffold-based cheminformatics approach for polypharmacological drug design.
Cell, 187, 2024
|
|
8JT8
 
 | | Crystal structure of 5-HT2AR in complex with (R)-IHCH-7179 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1-(4-fluorophenyl)-4-[(7R)-2,5,11-triazatetracyclo[7.6.1.0^2,7.0^12,16]hexadeca-1(15),9,12(16),13-tetraen-5-yl]butan-1-one, 5-hydroxytryptamine receptor 2A,Soluble cytochrome b562, ... | | Authors: | Chen, Z, Fan, L, Wang, S. | | Deposit date: | 2023-06-21 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Flexible scaffold-based cheminformatics approach for polypharmacological drug design.
Cell, 187, 2024
|
|
7UKL
 
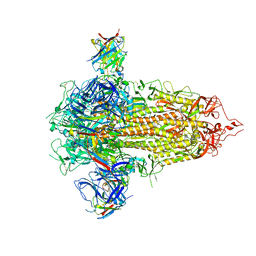 | |
7KJZ
 
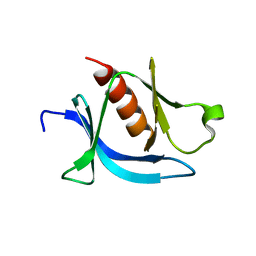 | | crystal structure of PLEKHA7 PH domain biding inositol-tetraphosphate | | Descriptor: | 1,2-ETHANEDIOL, INOSITOL-(1,3,4,5)-TETRAKISPHOSPHATE, Pleckstrin homology domain-containing family A member 7 | | Authors: | Marassi, F.M, Aleshin, A.E, Liddington, R.C. | | Deposit date: | 2020-10-26 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structural basis for the association of PLEKHA7 with membrane-embedded phosphatidylinositol lipids.
Structure, 29, 2021
|
|
7KJO
 
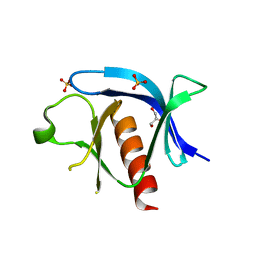 | |
7KK7
 
 | | crystal structure of ligand-free PLEKHA7 PH domain | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Pleckstrin homology domain-containing family A member 7 | | Authors: | Marassi, F.M, Aleshin, A.E, Liddington, R.C. | | Deposit date: | 2020-10-27 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for the association of PLEKHA7 with membrane-embedded phosphatidylinositol lipids.
Structure, 29, 2021
|
|
7L57
 
 | | Cryo-EM structure of the SARS-CoV-2 spike glycoprotein bound to Fab 2-15 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rapp, M, Shapiro, L. | | Deposit date: | 2020-12-21 | | Release date: | 2021-04-14 | | Last modified: | 2021-04-21 | | Method: | ELECTRON MICROSCOPY (5.87 Å) | | Cite: | Modular basis for potent SARS-CoV-2 neutralization by a prevalent VH1-2-derived antibody class.
Cell Rep, 35, 2021
|
|
7L56
 
 | | Cryo-EM structure of the SARS-CoV-2 spike glycoprotein bound to Fab 2-43 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab 2-43 variable domain heavy chain, ... | | Authors: | Rapp, M, Shapiro, L. | | Deposit date: | 2020-12-21 | | Release date: | 2021-04-14 | | Last modified: | 2023-04-05 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Modular basis for potent SARS-CoV-2 neutralization by a prevalent VH1-2-derived antibody class.
Cell Rep, 35, 2021
|
|
7L58
 
 | | Cryo-EM structure of the SARS-CoV-2 spike glycoprotein bound to Fab H4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab H4 variable domain heavy chain, ... | | Authors: | Rapp, M, Shapiro, L. | | Deposit date: | 2020-12-21 | | Release date: | 2021-04-14 | | Last modified: | 2021-04-21 | | Method: | ELECTRON MICROSCOPY (5.07 Å) | | Cite: | Modular basis for potent SARS-CoV-2 neutralization by a prevalent VH1-2-derived antibody class.
Cell Rep, 35, 2021
|
|
7CHS
 
 | | Crystal structure of SARS-CoV-2 antibody P22A-1D1 with RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, antibody P22A-1D1 heavy chain, ... | | Authors: | Wang, X, Zhang, L, Ge, J, Wang, R, Zhang, Q. | | Deposit date: | 2020-07-06 | | Release date: | 2021-05-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Potent and protective IGHV3-53/3-66 public antibodies and their shared escape mutant on the spike of SARS-CoV-2.
Nat Commun, 12, 2021
|
|
7CHO
 
 | | Crystal structure of SARS-CoV-2 antibody P5A-1D2 with RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, antibody P5A-1D2 heavy chain, ... | | Authors: | Wang, X, Zhang, L, Ge, J, Wang, R, Zhang, Q. | | Deposit date: | 2020-07-06 | | Release date: | 2021-05-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.561 Å) | | Cite: | Potent and protective IGHV3-53/3-66 public antibodies and their shared escape mutant on the spike of SARS-CoV-2.
Nat Commun, 12, 2021
|
|
7M8K
 
 | | Cryo-EM structure of Brazil (P.1) SARS-CoV-2 spike glycoprotein variant in the prefusion state (1 RBD up) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Casner, R.G, Cerutti, G, Shapiro, L, Ho, D.D. | | Deposit date: | 2021-03-29 | | Release date: | 2021-05-05 | | Last modified: | 2021-05-26 | | Method: | ELECTRON MICROSCOPY | | Cite: | Increased resistance of SARS-CoV-2 variant P.1 to antibody neutralization.
Cell Host Microbe, 29, 2021
|
|
7CHP
 
 | | Crystal structure of SARS-CoV-2 antibody P5A-3C8 with RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, antibody P5A-3C8 heavy chain, ... | | Authors: | Wang, X, Zhang, L, Ge, J, Wang, R, Zhang, Q. | | Deposit date: | 2020-07-06 | | Release date: | 2021-05-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.357 Å) | | Cite: | Potent and protective IGHV3-53/3-66 public antibodies and their shared escape mutant on the spike of SARS-CoV-2.
Nat Commun, 12, 2021
|
|
3SD8
 
 | | Crystal structure of Ara-FHNA decamer DNA | | Descriptor: | 5'-D(*GP*CP*GP*TP*AP*(F4H)P*AP*CP*GP*C)-3', STRONTIUM ION | | Authors: | Egli, M, Pallan, P.S. | | Deposit date: | 2011-06-08 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structural Origins of Opposite Effects on Stability by Axial and Equatorial 3'-Fluoro Modifications of Hexitol Nucleic Acid (HNA)
To be Published
|
|
5H05
 
 | | Crystal structure of AmyP E221Q in complex with MALTOTRIOSE | | Descriptor: | AmyP, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | He, C, Liu, Y. | | Deposit date: | 2016-10-03 | | Release date: | 2017-08-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of a raw-starch-degrading bacterial alpha-amylase belonging to subfamily 37 of the glycoside hydrolase family GH13
Sci Rep, 7, 2017
|
|
7XP6
 
 | | Cryo-EM structure of a class T GPCR in active state | | Descriptor: | Endoglucanase H,Taste receptor type 2 member 46,Endoglucanase H,Taste receptor type 2 member 46,Bitter taste receptor T2R46, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liu, Z.J, Hua, T, Xu, W.X, Wu, L.J. | | Deposit date: | 2022-05-03 | | Release date: | 2022-10-12 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structural basis for strychnine activation of human bitter taste receptor TAS2R46.
Science, 377, 2022
|
|
7XP4
 
 | | Cryo-EM structure of a class T GPCR in apo state | | Descriptor: | Endoglucanase H,Taste receptor type 2 member 46,Endoglucanase H,Taste receptor type 2 member 46,Bitter taste receptor T2R46, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liu, Z.J, Hua, T, Xu, W.X, Wu, L.J. | | Deposit date: | 2022-05-03 | | Release date: | 2022-10-12 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structural basis for strychnine activation of human bitter taste receptor TAS2R46.
Science, 377, 2022
|
|
