8HLM
 
 | | Crystal structure of p53/BCL2 fusion complex (complex 2) | | Descriptor: | Apoptosis regulator Bcl-2, Cellular tumor antigen p53, ZINC ION | | Authors: | Guo, M, Wang, H, Wei, H, Chen, Y. | | Deposit date: | 2022-11-30 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.522 Å) | | Cite: | Structures of p53/BCL-2 complex suggest a mechanism for p53 to antagonize BCL-2 activity.
Nat Commun, 14, 2023
|
|
8HLL
 
 | | Crystal structure of p53/BCL2 fusion complex (complex 1) | | Descriptor: | Apoptosis regulator Bcl-2, Cellular tumor antigen p53, ZINC ION | | Authors: | Wei, H, Guo, M, Wang, H, Chen, Y. | | Deposit date: | 2022-11-30 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structures of p53/BCL-2 complex suggest a mechanism for p53 to antagonize BCL-2 activity.
Nat Commun, 14, 2023
|
|
8HLN
 
 | | Crystal structure of p53/BCL2 fusion complex(complex3) | | Descriptor: | Apoptosis regulator Bcl-2, Cellular tumor antigen p53, ZINC ION | | Authors: | Guo, M, Wei, H, Wang, H, Chen, Y. | | Deposit date: | 2022-11-30 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.354 Å) | | Cite: | Structures of p53/BCL-2 complex suggest a mechanism for p53 to antagonize BCL-2 activity.
Nat Commun, 14, 2023
|
|
8HC2
 
 | | SARS-CoV-2 Omicron BA.1 spike trimer (6P) in complex with 1 YB9-258 Fab (1 RBD up) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of YB9-258 Fab, ... | | Authors: | Liu, B, Gao, X, Chen, Q, Li, Z, Su, M, He, J, Xiong, X. | | Deposit date: | 2022-11-01 | | Release date: | 2023-01-25 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (6.21 Å) | | Cite: | Somatically hypermutated antibodies isolated from SARS-CoV-2 Delta infected patients cross-neutralize heterologous variants.
Nat Commun, 14, 2023
|
|
8HCU
 
 | | Crystal structure of BCOR/PCGF1/KDM2B complex | | Descriptor: | ACETIC ACID, BCL-6 corepressor, Polycomb group RING finger protein 1, ... | | Authors: | Shen, F, Chen, R, Xu, J, Liu, J. | | Deposit date: | 2022-11-03 | | Release date: | 2023-11-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Calcium modulates the tethering of BCOR-PRC1.1 enzymatic core to KDM2B via liquid-liquid phase separation.
Commun Biol, 7, 2024
|
|
8HIH
 
 | | Cryo-EM structure of Mycobacterium tuberculosis transcription initiation complex with transcription factor GlnR | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Lin, W, Shi, J, Xu, J.C. | | Deposit date: | 2022-11-20 | | Release date: | 2023-06-07 | | Last modified: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Structural insights into the transcription activation mechanism of the global regulator GlnR from actinobacteria.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8IMX
 
 | | Cryo-EM structure of GPI-T with a chimeric GPI-anchored protein | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Xu, Y, Li, T, Qu, Q, Li, D. | | Deposit date: | 2023-03-07 | | Release date: | 2023-08-16 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structures of liganded glycosylphosphatidylinositol transamidase illuminate GPI-AP biogenesis.
Nat Commun, 14, 2023
|
|
8IS5
 
 | |
8IS4
 
 | | Structure of an Isocytosine specific deaminase Vcz in complexed with 5-FU | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5-FLUOROURACIL, GLYCEROL, ... | | Authors: | Guo, W.T, Li, X.J, Wu, B.X. | | Deposit date: | 2023-03-20 | | Release date: | 2023-08-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural characterization of an isocytosine-specific deaminase VCZ reveals its application potential in the anti-cancer therapy.
Iscience, 26, 2023
|
|
8IMY
 
 | | Cryo-EM structure of GPI-T (inactive mutant) with GPI and proULBP2, a proprotein substrate | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, T, Xu, Y, Qu, Q, Li, D. | | Deposit date: | 2023-03-07 | | Release date: | 2023-08-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Structures of liganded glycosylphosphatidylinositol transamidase illuminate GPI-AP biogenesis.
Nat Commun, 14, 2023
|
|
7YE9
 
 | | SARS-CoV-2 Spike (6P) in complex with 3 R1-32 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of R1-32 Fab, ... | | Authors: | Liu, B, Gao, X, Li, Z, Chen, X, He, J, Chen, L, Xiong, X. | | Deposit date: | 2022-07-05 | | Release date: | 2022-08-24 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (4.17 Å) | | Cite: | SARS-CoV-2 Delta and Omicron variants evade population antibody response by mutations in a single spike epitope.
Nat Microbiol, 7, 2022
|
|
7YDY
 
 | | SARS-CoV-2 Spike (6P) in complex with 1 R1-32 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of R1-32 Fab, ... | | Authors: | Liu, B, Gao, X, Li, Z, Chen, X, He, J, Chen, L, Xiong, X. | | Deposit date: | 2022-07-04 | | Release date: | 2022-08-24 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (4.75 Å) | | Cite: | SARS-CoV-2 Delta and Omicron variants evade population antibody response by mutations in a single spike epitope.
Nat Microbiol, 7, 2022
|
|
7YEG
 
 | | SARS-CoV-2 Spike (6P) in complex with 3 R1-32 Fabs and 3 ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Liu, B, Gao, X, Li, Z, Chen, X, He, J, Chen, L, Xiong, X. | | Deposit date: | 2022-07-05 | | Release date: | 2022-08-24 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | SARS-CoV-2 Delta and Omicron variants evade population antibody response by mutations in a single spike epitope.
Nat Microbiol, 7, 2022
|
|
7YDI
 
 | | SARS-CoV-2 Spike (6P) in complex with 3 R1-32 Fabs and 3 ACE2, focused refinement of RBD region | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of R1-32, Light chain of R1-32, ... | | Authors: | Liu, B, Gao, X, Li, Z, Chen, X, He, J, Chen, L, Xiong, X. | | Deposit date: | 2022-07-04 | | Release date: | 2022-08-24 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.98 Å) | | Cite: | SARS-CoV-2 Delta and Omicron variants evade population antibody response by mutations in a single spike epitope.
Nat Microbiol, 7, 2022
|
|
7WDM
 
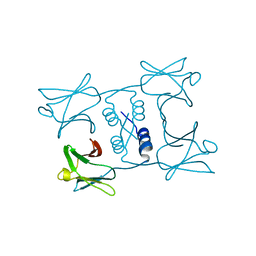 | | Fungal immunomodulatory protein FIP-gmi | | Descriptor: | Immunomodulatory protein | | Authors: | Liu, Y, Bastiaan-Net, S, Zhang, Y, Hoppenbrouwers, T, Xie, Y, Wang, Y, Wei, X, Du, G, Zhang, H, Imam, K.M.S.U, Wichers, H.J, Li, Z. | | Deposit date: | 2021-12-22 | | Release date: | 2022-12-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.123 Å) | | Cite: | Linking the thermostability of FIP-nha (Nectria haematococca) to its structural properties
To Be Published
|
|
7YDG
 
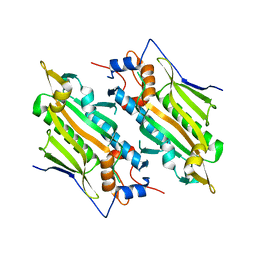 | | Crystal structure of human SARS2 catalytic domain with a disease related mutation | | Descriptor: | Serine--tRNA ligase, mitochondrial | | Authors: | Wu, S, Li, P, Zhou, X.L, Fang, P. | | Deposit date: | 2022-07-04 | | Release date: | 2022-11-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Selective degradation of tRNASer(AGY) is the primary driver for mitochondrial seryl-tRNA synthetase-related disease.
Nucleic Acids Res., 50, 2022
|
|
7YDF
 
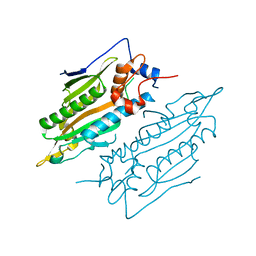 | | Crystal structure of human SARS2 catalytic domain | | Descriptor: | Serine--tRNA ligase, mitochondrial | | Authors: | Wu, S, Li, P, Zhou, X.L, Fang, P. | | Deposit date: | 2022-07-04 | | Release date: | 2022-11-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Selective degradation of tRNASer(AGY) is the primary driver for mitochondrial seryl-tRNA synthetase-related disease.
Nucleic Acids Res., 50, 2022
|
|
7YE5
 
 | | SARS-CoV-2 Spike (6P) in complex with 2 R1-32 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of R1-32 Fab, ... | | Authors: | Liu, B, Gao, X, Li, Z, Chen, X, He, J, Chen, L, Xiong, X. | | Deposit date: | 2022-07-05 | | Release date: | 2022-08-24 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (6.75 Å) | | Cite: | SARS-CoV-2 Delta and Omicron variants evade population antibody response by mutations in a single spike epitope.
Nat Microbiol, 7, 2022
|
|
1CC0
 
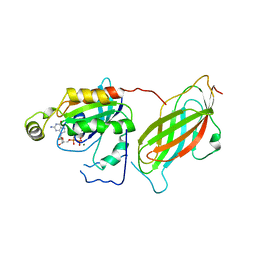 | | CRYSTAL STRUCTURE OF THE RHOA.GDP-RHOGDI COMPLEX | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, rho GDP dissociation inhibitor alpha, ... | | Authors: | Longenecker, K.L, Read, P, Derewenda, U, Dauter, Z, Garrard, S, Walker, L, Somlyo, A.V, Somlyo, A.P, Nakamoto, R.K, Derewenda, Z.S. | | Deposit date: | 1999-03-03 | | Release date: | 2000-01-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (5 Å) | | Cite: | How RhoGDI binds Rho.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
6H8P
 
 | | JMJD2A/ KDM4A COMPLEXED WITH NI(II), NOG AND Histone H1.4(18-32)K26me3 peptide (15-mer) | | Descriptor: | CHLORIDE ION, GLYCEROL, Histone H1.4, ... | | Authors: | Chowdhury, R, Walport, L.J, Schofield, C.J. | | Deposit date: | 2018-08-03 | | Release date: | 2018-08-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.983 Å) | | Cite: | Mechanistic and structural studies of KDM-catalysed demethylation of histone 1 isotype 4 at lysine 26.
FEBS Lett., 592, 2018
|
|
7BSX
 
 | | SDR protein NapW-NADP | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Short chain dehydrogenase | | Authors: | Wen, W.H, Tang, G.L. | | Deposit date: | 2020-03-31 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Reductive inactivation of the hemiaminal pharmacophore for resistance against tetrahydroisoquinoline antibiotics.
Nat Commun, 12, 2021
|
|
7BTM
 
 | | SDR protein/resistance protein NapW | | Descriptor: | Short chain dehydrogenase | | Authors: | Wen, W.H, Tang, G.L. | | Deposit date: | 2020-04-02 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.08311653 Å) | | Cite: | Reductive inactivation of the hemiaminal pharmacophore for resistance against tetrahydroisoquinoline antibiotics.
Nat Commun, 12, 2021
|
|
6IVE
 
 | |
8HK0
 
 | | Crystal structure of Fic32-33 complex from Streptomyces ficellus NRRL 8067 | | Descriptor: | Acyl-CoA dehydrogenase, Dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Cheng, Y, Qiao, H, Liu, W, Fang, P. | | Deposit date: | 2022-11-24 | | Release date: | 2023-04-26 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Oxidase Heterotetramer Completes 1-Azabicyclo[3.1.0]hexane Formation with the Association of a Nonribosomal Peptide Synthetase.
J.Am.Chem.Soc., 145, 2023
|
|
7C7U
 
 | | Biofilm associated protein - BSP domain | | Descriptor: | Biofilm-associated surface protein, CALCIUM ION | | Authors: | Ma, J.F, Xu, Z.H, Zhang, Y.K, Cheng, X, Fan, S.L, Wang, J.W, Fang, X.Y. | | Deposit date: | 2020-05-26 | | Release date: | 2021-05-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural mechanism for modulation of functional amyloid and biofilm formation by Staphylococcal Bap protein switch.
Embo J., 40, 2021
|
|
