1IDE
 
 | | ISOCITRATE DEHYDROGENASE Y160F MUTANT STEADY-STATE INTERMEDIATE COMPLEX (LAUE DETERMINATION) | | Descriptor: | ISOCITRATE DEHYDROGENASE, ISOCITRIC ACID, MAGNESIUM ION, ... | | Authors: | Bolduc, J.M, Dyer, D.H, Scott, W.G, Singer, P, Sweet, R.M, Koshland Junior, D.E, Stoddard, B.L. | | Deposit date: | 1995-01-18 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mutagenesis and Laue structures of enzyme intermediates: isocitrate dehydrogenase.
Science, 268, 1995
|
|
3KL4
 
 | | Recognition of a signal peptide by the signal recognition particle | | Descriptor: | Signal peptide of yeast dipeptidyl aminopeptidase B, Signal recognition 54 kDa protein | | Authors: | Janda, C.Y, Nagai, K, Li, J, Oubridge, C. | | Deposit date: | 2009-11-06 | | Release date: | 2010-03-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Recognition of a signal peptide by the signal recognition particle.
Nature, 465, 2010
|
|
1TIS
 
 | |
1IDD
 
 | | ISOCITRATE DEHYDROGENASE Y160F MUTANT APO ENZYME | | Descriptor: | ISOCITRATE DEHYDROGENASE | | Authors: | Lee, M.E, Dyer, D.H, Klein, O.D, Bolduc, J.M, Stoddard, B.L, Koshland Junior, D.E. | | Deposit date: | 1995-01-18 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mutagenesis and Laue structures of enzyme intermediates: isocitrate dehydrogenase.
Science, 268, 1995
|
|
3UWL
 
 | | Crystal structure of Enteroccocus faecalis thymidylate synthase (EfTS) in complex with 5-formyl tetrahydrofolate | | Descriptor: | 1,2-ETHANEDIOL, N-[4-({[(6S)-2-amino-5-formyl-4-oxo-3,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)benzoyl]-L-glutamic acid, SULFATE ION, ... | | Authors: | Pozzi, C, Catalano, A, Cortesi, D, Luciani, R, Ferrari, S, Fritz, T, Costi, M.P, Mangani, S. | | Deposit date: | 2011-12-02 | | Release date: | 2012-08-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | The structure of Enterococcus faecalis thymidylate synthase provides clues about folate bacterial metabolism.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3B7D
 
 | |
1THY
 
 | |
8SBE
 
 | | Structure of the rat vesicular glutamate transporter 2 determined by single-particle Cryo-EM | | Descriptor: | Vesicular glutamate transporter 2 | | Authors: | Li, F, Finer-Moore, J, Eriksen, J, Cheng, Y, Edwards, R, Stroud, R. | | Deposit date: | 2023-04-03 | | Release date: | 2023-05-03 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Ion transport and regulation in a synaptic vesicle glutamate transporter.
Science, 368, 2020
|
|
8VKJ
 
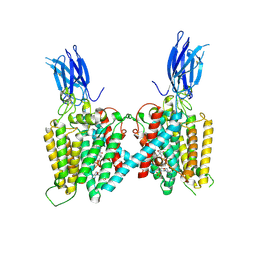 | | Cryo-EM structure of human HGSNAT bound with Acetyl-CoA | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYL COENZYME *A, ... | | Authors: | Li, F, Zhao, B. | | Deposit date: | 2024-01-09 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Structural and mechanistic insights into a lysosomal membrane enzyme HGSNAT involved in Sanfilippo syndrome.
Nat Commun, 15, 2024
|
|
8VLV
 
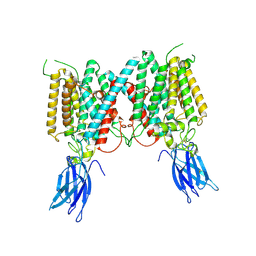 | | Cryo-EM structure of human HGSNAT in inactive state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heparan-alpha-glucosaminide N-acetyltransferase | | Authors: | Li, F. | | Deposit date: | 2024-01-12 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Structural and mechanistic insights into a lysosomal membrane enzyme HGSNAT involved in Sanfilippo syndrome.
Nat Commun, 15, 2024
|
|
8VLI
 
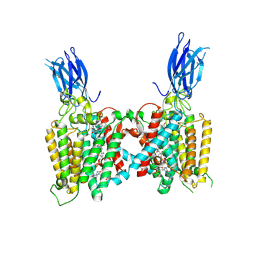 | | Cryo-EM structure of human HGSNAT bound with CoA and product analog | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-methyl-2-oxo-2H-1-benzopyran-7-yl 2-acetamido-2-deoxy-beta-D-glucopyranoside, ... | | Authors: | Li, F, Zhao, B. | | Deposit date: | 2024-01-11 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and mechanistic insights into a lysosomal membrane enzyme HGSNAT involved in Sanfilippo syndrome.
Nat Commun, 15, 2024
|
|
8VLG
 
 | | Cryo-EM structure of human HGSNAT bound with Acetyl-CoA and substrate analog | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-methyl-2-oxo-2H-1-benzopyran-7-yl 2-amino-2-deoxy-beta-D-glucopyranoside, ... | | Authors: | Li, F, Zhao, B. | | Deposit date: | 2024-01-11 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Structural and mechanistic insights into a lysosomal membrane enzyme HGSNAT involved in Sanfilippo syndrome.
Nat Commun, 15, 2024
|
|
8VLU
 
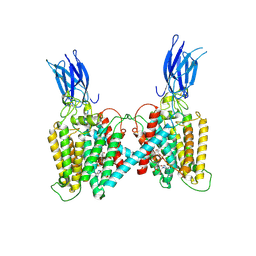 | | Cryo-EM structure of human HGSNAT bound with CoA | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COENZYME A, Heparan-alpha-glucosaminide N-acetyltransferase | | Authors: | Li, F, Zhao, B. | | Deposit date: | 2024-01-12 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Structural and mechanistic insights into a lysosomal membrane enzyme HGSNAT involved in Sanfilippo syndrome.
Nat Commun, 15, 2024
|
|
8VLY
 
 | | Cryo-EM structure of human HGSNAT in transition state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heparan-alpha-glucosaminide N-acetyltransferase | | Authors: | Li, F, Zhao, B. | | Deposit date: | 2024-01-12 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.61 Å) | | Cite: | Structural and mechanistic insights into a lysosomal membrane enzyme HGSNAT involved in Sanfilippo syndrome.
Nat Commun, 15, 2024
|
|
3SDJ
 
 | | Structure of RNase-inactive point mutant of oligomeric kinase/RNase Ire1 | | Descriptor: | N~2~-1H-benzimidazol-5-yl-N~4~-(3-cyclopropyl-1H-pyrazol-5-yl)pyrimidine-2,4-diamine, Serine/threonine-protein kinase/endoribonuclease IRE1 | | Authors: | Korennykh, A, Korostelev, A, Egea, P, Finer-Moore, J, Zhang, C, Stroud, R, Shokat, K, Walter, P. | | Deposit date: | 2011-06-09 | | Release date: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Structural and functional basis for RNA cleavage by Ire1.
Bmc Biol., 9, 2011
|
|
1CW7
 
 | |
1CW1
 
 | |
1BL5
 
 | | ISOCITRATE DEHYDROGENASE FROM E. COLI SINGLE TURNOVER LAUE STRUCTURE OF RATE-LIMITED PRODUCT COMPLEX, 10 MSEC TIME RESOLUTION | | Descriptor: | 2-OXOGLUTARIC ACID, ISOCITRATE DEHYDROGENASE, MAGNESIUM ION, ... | | Authors: | Stoddard, B.L, Cohen, B, Brubaker, M, Mesecar, A, Koshland Junior, D.E. | | Deposit date: | 1998-07-23 | | Release date: | 1999-05-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Millisecond Laue structures of an enzyme-product complex using photocaged substrate analogs.
Nat.Struct.Biol., 5, 1998
|
|
5HS1
 
 | | Saccharomyces cerevisiae CYP51 (Lanosterol 14-alpha demethylase) complexed with Voriconazole | | Descriptor: | Lanosterol 14-alpha demethylase, PROTOPORPHYRIN IX CONTAINING FE, Voriconazole | | Authors: | Sabherwal, M, Sagatova, A, Keniya, M.V, Wilson, R.K, Tyndall, J.D.A, Monk, B.C. | | Deposit date: | 2016-01-24 | | Release date: | 2016-06-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Triazole resistance mediated by mutations of a conserved active site tyrosine in fungal lanosterol 14 alpha-demethylase.
Sci Rep, 6, 2016
|
|
1GRO
 
 | |
1GRP
 
 | |
4WMZ
 
 | | S. cerevisiae CYP51 complexed with fluconazole in the active site | | Descriptor: | 2-(2,4-DIFLUOROPHENYL)-1,3-DI(1H-1,2,4-TRIAZOL-1-YL)PROPAN-2-OL, Lanosterol 14-alpha demethylase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sagatova, A, Keniya, M.V, Wilson, R, Wilbanks, S.M, Monk, B.C, Tyndall, J.D.A. | | Deposit date: | 2014-10-09 | | Release date: | 2015-07-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Insights into Binding of the Antifungal Drug Fluconazole to Saccharomyces cerevisiae Lanosterol 14 alpha-Demethylase.
Antimicrob.Agents Chemother., 59, 2015
|
|
1CW4
 
 | |
1CII
 
 | |
5JLC
 
 | | Structure of CYP51 from the pathogen Candida glabrata | | Descriptor: | 2-[(2R)-butan-2-yl]-4-{4-[4-(4-{[(2R,4S)-2-(2,4-dichlorophenyl)-2-(1H-1,2,4-triazol-1-ylmethyl)-1,3-dioxolan-4-yl]methoxy}phenyl)piperazin-1-yl]phenyl}-2,4-dihydro-3H-1,2,4-triazol-3-one, CHLORIDE ION, Lanosterol 14-alpha demethylase, ... | | Authors: | Keniya, M.V, Sabherwal, M, Wilson, R.K, Sagatova, A.A, Tyndall, J.D.A, Monk, B.C. | | Deposit date: | 2016-04-26 | | Release date: | 2016-05-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of Full-Length Lanosterol 14 alpha-Demethylases of Prominent Fungal Pathogens Candida albicans and Candida glabrata Provide Tools for Antifungal Discovery.
Antimicrob.Agents Chemother., 62, 2018
|
|
