7X9U
 
 | | Type-II KH motif of human mitochondrial RbfA | | Descriptor: | Putative ribosome-binding factor A, mitochondrial | | Authors: | Kuwasako, K, Suzuki, S, Furue, M, Takizawa, M, Takahashi, M, Tsuda, K, Nagata, T, Watanabe, S, Tanaka, A, Kobayashi, N, Kigawa, T, Guntert, P, Shirouzu, M, Yokoyama, S, Muto, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2022-03-16 | | Release date: | 2023-01-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | 1 H, 13 C, and 15 N resonance assignments and solution structures of the KH domain of human ribosome binding factor A, mtRbfA, involved in mitochondrial ribosome biogenesis.
Biomol.Nmr Assign., 16, 2022
|
|
2RPP
 
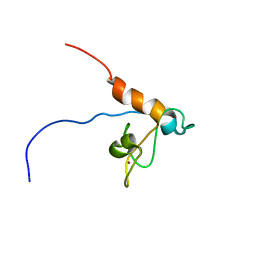 | | Solution structure of Tandem zinc finger domain 12 in Muscleblind-like protein 2 | | Descriptor: | Muscleblind-like protein 2, ZINC ION | | Authors: | Abe, C, Dang, W, Tsuda, K, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-06-24 | | Release date: | 2009-05-12 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the RNA binding domain in the human muscleblind-like protein 2
Protein Sci., 18, 2009
|
|
2YTZ
 
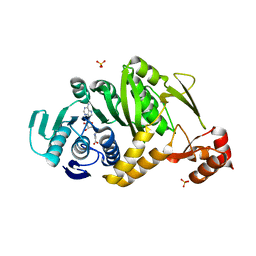 | | Complex structure of Trm1 from Pyrococcus horikoshii with S-adenosyl-L-Homocystein in the orthorhombic crystal-lattice | | Descriptor: | N(2),N(2)-dimethylguanosine tRNA methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION | | Authors: | Ihsanawati, Shirouzu, M, Bessho, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-05 | | Release date: | 2007-10-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal Structure of tRNA N(2),N(2)-Guanosine Dimethyltransferase Trm1 from Pyrococcus horikoshii
J.Mol.Biol., 383, 2008
|
|
2YV8
 
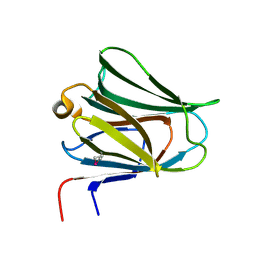 | | Crystal structure of N-terminal domain of human galectin-8 | | Descriptor: | Galectin-8 variant | | Authors: | Kishishita, S, Nishino, A, Murayama, K, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-10 | | Release date: | 2008-04-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structure of N-terminal domain of human galectin-8
To be Published
|
|
2YV6
 
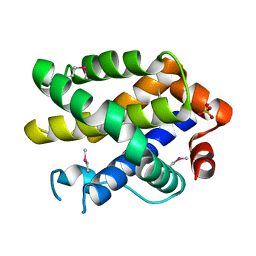 | | Crystal structure of human Bcl-2 family protein Bak | | Descriptor: | Bcl-2 homologous antagonist/killer, SULFATE ION | | Authors: | Wang, H, Kishishita, S, Murayama, K, Takemoto, C, Terada, T, Shirouzu, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-09 | | Release date: | 2008-04-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Novel dimerization mode of the human Bcl-2 family protein Bak, a mitochondrial apoptosis regulator.
J.Struct.Biol., 166, 2009
|
|
2CZG
 
 | | Crystal structure of Probable phosphoribosylglycinamide formyl transferase (PH0318) from Pyrococcus horikoshii OT3 | | Descriptor: | GLYCEROL, SULFATE ION, phosphoribosylglycinamide formyl transferase | | Authors: | Yoshikawa, S, Arai, R, Kamo-Uchikubo, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-13 | | Release date: | 2006-01-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of Probable phosphoribosylglycinamide formyl transferase (PH0318) from Pyrococcus horikoshii OT3
To be Published
|
|
2ZDY
 
 | | Inhibitor-bound structures of human pyruvate dehydrogenase kinase 4 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Kawamoto, M, Shiromizu, I, Kukimoto-Niino, M, Tokmakov, A, Terada, T, Shirouzu, M, Matsusue, T, Yokoyama, S. | | Deposit date: | 2007-12-01 | | Release date: | 2008-12-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Inhibitor-bound structures of human pyruvate dehydrogenase kinase 4.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
2ZET
 
 | | Crystal structure of the small GTPase Rab27B complexed with the Slp homology domain of Slac2-a/melanophilin | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Melanophilin, ... | | Authors: | Kukimoto-Niino, M, Sakamoto, A, Kanno, E, Hanawa-Suetsugu, K, Terada, T, Shirouzu, M, Fukuda, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-12-17 | | Release date: | 2008-09-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for the exclusive specificity of Slac2-a/melanophilin for the Rab27 GTPases.
Structure, 16, 2008
|
|
2ZXV
 
 | | Crystal structure of putative acetyltransferase from T. thermophilus HB8 | | Descriptor: | Putative uncharacterized protein TTHA1799 | | Authors: | Murayama, K, Kato-Murayama, M, Terada, T, Kuramitsu, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-01-08 | | Release date: | 2009-02-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Genetic Encoding of 3-Iodo-l-Tyrosine in Escherichia coli for Single-Wavelength Anomalous Dispersion Phasing in Protein Crystallography
Structure, 17, 2009
|
|
3A3U
 
 | | Crystal structure of MqnD (TTHA1568), a menaquinone biosynthetic enzyme from Thermus thermophilus HB8 | | Descriptor: | 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, L(+)-TARTARIC ACID, Menaquinone biosynthetic enzyme, ... | | Authors: | Arai, R, Nishino, A, Nagano, K, Uchikubo-Kamo, T, Katsura, K, Nishimoto, M, Toyama, M, Terada, T, Kuramitsu, S, Murayama, K, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-06-20 | | Release date: | 2009-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of MqnD (TTHA1568), a menaquinone biosynthetic enzyme from Thermus thermophilus HB8.
J.Struct.Biol., 168, 2009
|
|
3VJN
 
 | | Crystal structure of the mutated EGFR kinase domain (G719S/T790M) in complex with AMPPNP. | | Descriptor: | Epidermal growth factor receptor, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Yoshikawa, S, Kukimoto-Niino, M, Shirouzu, M, Semba, K, Yamamoto, T, Yokoyama, S. | | Deposit date: | 2011-10-27 | | Release date: | 2012-03-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structural basis for the altered drug sensitivities of non-small cell lung cancer-associated mutants of human epidermal growth factor receptor.
Oncogene, 32, 2013
|
|
1WQU
 
 | | Solution structure of the human FES SH2 domain | | Descriptor: | Proto-oncogene tyrosine-protein kinase FES/FPS | | Authors: | Scott, A, Pantoja-Uceda, D, Koshiba, S, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Tanaka, A, Sugano, S, Yokoyama, S, Guntert, P, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-10-02 | | Release date: | 2005-06-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Src homology 2 domain from the human feline sarcoma oncogene Fes
J.Biomol.NMR, 31, 2005
|
|
7VGU
 
 | | Time-resolved serial femtosecond crystallography structure of light-driven chloride ion-pumping rhodopsin, NM-R3 : structure obtained 1 msec after photoexcitation with bromide ion | | Descriptor: | BROMIDE ION, Chloride pumping rhodopsin, DECANE, ... | | Authors: | Hosaka, T, Nango, E, Nakane, T, Luo, F, Kimura-Someya, T, Shirouzu, M. | | Deposit date: | 2021-09-18 | | Release date: | 2022-02-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Conformational alterations in unidirectional ion transport of a light-driven chloride pump revealed using X-ray free electron lasers.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6A5L
 
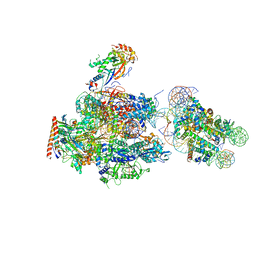 | | RNA polymerase II elongation complex stalled at SHL(-1) of the nucleosome, with foreign DNA | | Descriptor: | DNA (198-MER), DNA (42-MER), DNA-directed RNA polymerase subunit, ... | | Authors: | Kujirai, T, Ehara, H, Fujino, Y, Shirouzu, M, Sekine, S, Kurumizaka, H. | | Deposit date: | 2018-06-24 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Structural basis of the nucleosome transition during RNA polymerase II passage.
Science, 362, 2018
|
|
7VGT
 
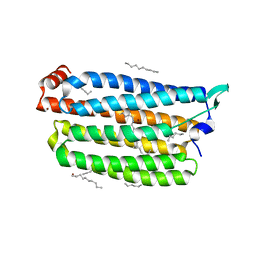 | | Time-resolved serial femtosecond crystallography structure of light-driven chloride ion-pumping rhodopsin, NM-R3: resting state structure with bromide ion | | Descriptor: | BROMIDE ION, Chloride pumping rhodopsin, DECANE, ... | | Authors: | Hosaka, T, Nango, E, Nakane, T, Luo, F, Kimura-Someya, T, Shirouzu, M. | | Deposit date: | 2021-09-18 | | Release date: | 2022-02-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Conformational alterations in unidirectional ion transport of a light-driven chloride pump revealed using X-ray free electron lasers.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7VGV
 
 | | Anion free form of light-driven chloride ion-pumping rhodopsin, NM-R3, structure determined by serial femtosecond crystallography at SACLA | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, HEXADECANE, ... | | Authors: | Hosaka, T, Nango, E, Nakane, T, Luo, F, Kimura-Someya, T, Shirouzu, M. | | Deposit date: | 2021-09-18 | | Release date: | 2022-02-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Conformational alterations in unidirectional ion transport of a light-driven chloride pump revealed using X-ray free electron lasers.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6A5R
 
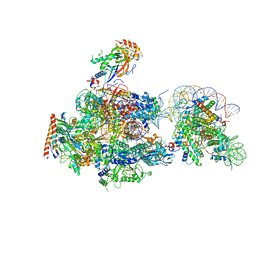 | | RNA polymerase II elongation complex stalled at SHL(-2) of the nucleosome | | Descriptor: | DNA (198-MER), DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Kujirai, T, Ehara, H, Fujino, Y, Shirouzu, M, Sekine, S, Kurumizaka, H. | | Deposit date: | 2018-06-25 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (8.7 Å) | | Cite: | Structural basis of the nucleosome transition during RNA polymerase II passage.
Science, 362, 2018
|
|
3VHL
 
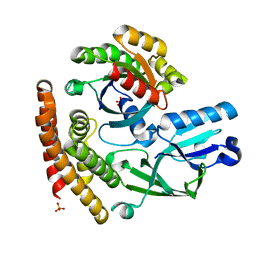 | | Crystal structure of the DHR-2 domain of DOCK8 in complex with Cdc42 (T17N mutant) | | Descriptor: | Cell division control protein 42 homolog, Dedicator of cytokinesis protein 8, PHOSPHATE ION | | Authors: | Hanawa-Suetsugu, K, Kukimoto-Niino, M, Nishizak, T, Terada, T, Shirouzu, M, Fukui, Y, Yokoyama, S. | | Deposit date: | 2011-08-26 | | Release date: | 2012-06-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.085 Å) | | Cite: | DOCK8 is a Cdc42 activator critical for interstitial dendritic cell migration during immune responses.
Blood, 119, 2012
|
|
6A5P
 
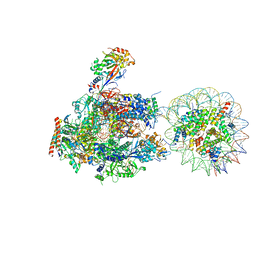 | | RNA polymerase II elongation complex stalled at SHL(-5) of the nucleosome | | Descriptor: | DNA (198-MER), DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Kujirai, T, Ehara, H, Fujino, Y, Shirouzu, M, Sekine, S, Kurumizaka, H. | | Deposit date: | 2018-06-25 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Structural basis of the nucleosome transition during RNA polymerase II passage.
Science, 362, 2018
|
|
6A5O
 
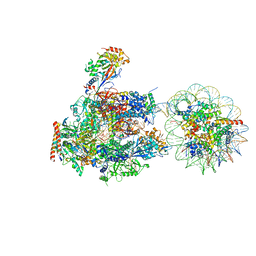 | | RNA polymerase II elongation complex stalled at SHL(-6) of the nucleosome | | Descriptor: | DNA (198-MER), DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Kujirai, T, Ehara, H, Fujino, Y, Shirouzu, M, Sekine, S, Kurumizaka, H. | | Deposit date: | 2018-06-25 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (9.9 Å) | | Cite: | Structural basis of the nucleosome transition during RNA polymerase II passage.
Science, 362, 2018
|
|
2ZKY
 
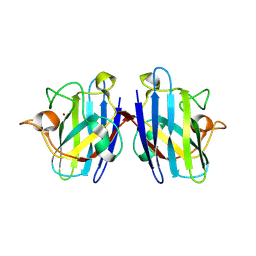 | | Crystal structure of human Cu-Zn superoxide dismutase mutant G93A | | Descriptor: | Superoxide dismutase [Cu-Zn], ZINC ION | | Authors: | Yoshikawa, S, Kukimoto-Niino, M, Ito, K, Shirouzu, M, Urushitani, M, Takahashi, R, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-03-31 | | Release date: | 2009-03-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of human Cu-Zn superoxide dismutase mutant G93A
To be Published
|
|
6ACR
 
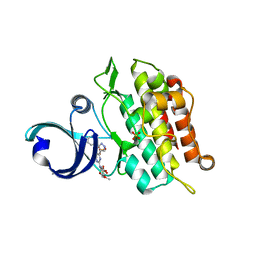 | | Crystal structure of human ALK2 kinase domain with R206H mutation in complex with RK-59638 | | Descriptor: | Activin receptor type-1, N-(4-methoxyphenyl)-4-[3-(pyridin-3-yl)-1H-pyrazol-4-yl]pyrimidin-2-amine, SULFATE ION | | Authors: | Sakai, N, Mishima-Tsumagari, C, Matsumoto, T, Shirouzu, M. | | Deposit date: | 2018-07-27 | | Release date: | 2019-03-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Bis-Heteroaryl Pyrazoles: Identification of Orally Bioavailable Inhibitors of Activin Receptor-Like Kinase-2 (R206H).
Chem. Pharm. Bull., 67, 2019
|
|
5ZAB
 
 | | Crystal structure of cf3-aequorin | | Descriptor: | (2S)-8-benzyl-2-hydroperoxy-6-(4-hydroxyphenyl)-2-{[4-(trifluoromethyl)phenyl]methyl}imidazo[1,2-a]pyrazin-3(2H)-one, Aequorin-2 | | Authors: | Inouye, S, Tomabechi, Y, Sekine, S.I, Shirouzu, M, Hosoya, T. | | Deposit date: | 2018-02-07 | | Release date: | 2018-06-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.147 Å) | | Cite: | Slow luminescence kinetics of semi-synthetic aequorin: expression, purification and structure determination of cf3-aequorin.
J. Biochem., 164, 2018
|
|
3UG1
 
 | | Crystal structure of the mutated EGFR kinase domain (G719S/T790M) in the apo form | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Epidermal growth factor receptor | | Authors: | Parker, L.J, Handa, N, Yoshikawa, S, Kukimoto-Niino, M, Shirouzu, M, Yokoyama, S. | | Deposit date: | 2011-11-02 | | Release date: | 2012-03-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis for the altered drug sensitivities of non-small cell lung cancer-associated mutants of human epidermal growth factor receptor
Oncogene, 32, 2013
|
|
2ZKW
 
 | | Crystal structure of human Cu-Zn superoxide dismutase mutant G85R in space group P21 | | Descriptor: | COPPER (I) ION, Superoxide dismutase [Cu-Zn], ZINC ION | | Authors: | Yoshikawa, S, Kukimoto-Niino, M, Ito, K, Chen, L, Fu, Z.Q, Chrzas, J, Wang, B.C, Shirouzu, M, Urushitani, M, Takahashi, R, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-03-31 | | Release date: | 2009-03-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of human Cu-Zn superoxide dismutase mutant G85R in space group P21
To be Published
|
|
