8Y5H
 
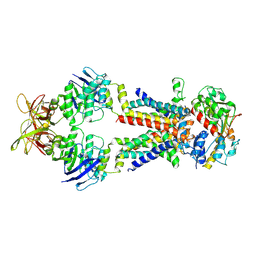 | |
8Y5F
 
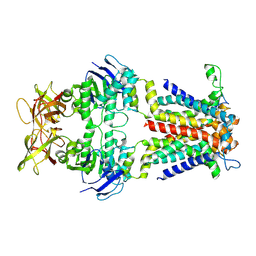 | | Cryo-EM structure of E.coli spermidine transporter PotABC | | Descriptor: | Spermidine/putrescine import ATP-binding protein PotA, Spermidine/putrescine transport system permease protein PotB, Spermidine/putrescine transport system permease protein PotC | | Authors: | Qiao, Z, Gao, Y.G. | | Deposit date: | 2024-01-31 | | Release date: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Structural insights into polyamine spermidine uptake by the ABC transporter PotD-PotABC.
Sci Adv, 10, 2024
|
|
8Y5G
 
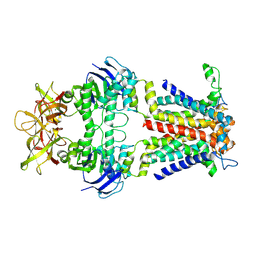 | |
8Y5I
 
 | |
6TN3
 
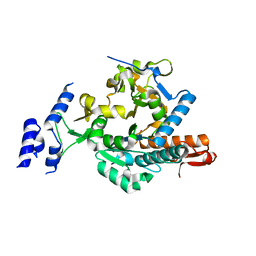 | |
1HKA
 
 | | 6-HYDROXYMETHYL-7,8-DIHYDROPTERIN PYROPHOSPHOKINASE | | Descriptor: | 6-HYDROXYMETHYL-7,8-DIHYDROPTERIN PYROPHOSPHOKINASE | | Authors: | Xiao, B, Shi, G, Chen, X, Yan, H, Ji, X. | | Deposit date: | 1998-09-29 | | Release date: | 1999-06-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase, a potential target for the development of novel antimicrobial agents.
Structure Fold.Des., 7, 1999
|
|
4LXZ
 
 | | Structure of Human HDAC2 in complex with SAHA (vorinostat) | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, CALCIUM ION, Histone deacetylase 2, ... | | Authors: | Fong, R, Lupardus, P.J. | | Deposit date: | 2013-07-30 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Histone Deacetylase (HDAC) Inhibitor Kinetic Rate Constants Correlate with Cellular Histone Acetylation but Not Transcription and Cell Viability.
J.Biol.Chem., 288, 2013
|
|
4UMI
 
 | |
3ZBT
 
 | | Ferredoxin-NADP Reductase Mutant with SER 59 Replaced by ALA (S59A) | | Descriptor: | FERREDOXIN-NADP REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Martinez-Julvez, M, Herguedas, B, Sanchez-Azqueta, A, Hervas, M, Navarro, J.A, Medina, M. | | Deposit date: | 2012-11-13 | | Release date: | 2013-11-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | A Hydrogen Bond Network in the Active Site of Anabaena Ferredoxin-Nadp(+) Reductase Modulates its Catalytic Efficiency.
Biochim.Biophys.Acta, 1837, 2013
|
|
8CR8
 
 | | human Interleukin-23 | | Descriptor: | Interleukin-12 subunit beta, Interleukin-23 subunit alpha, TERBIUM(III) ION, ... | | Authors: | Bloch, Y, Savvides, S.N. | | Deposit date: | 2023-03-08 | | Release date: | 2024-02-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of complete extracellular receptor assemblies mediated by IL-12 and IL-23.
Nat.Struct.Mol.Biol., 31, 2024
|
|
3ZBU
 
 | | Ferredoxin-NADP Reductase Mutant with SER 80 Replaced by ALA (S80A) | | Descriptor: | FERREDOXIN-NADP REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Martinez-Julvez, M, Herguedas, B, Sanchez-Azqueta, A, Hervas, M, Navarro, J.A, Medina, M. | | Deposit date: | 2012-11-13 | | Release date: | 2013-11-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | A Hydrogen Bond Network in the Active Site of Anabaena Ferredoxin-Nadp(+) Reductase Modulates its Catalytic Efficiency.
Biochim.Biophys.Acta, 1837, 2013
|
|
2Q5E
 
 | | Crystal structure of human carboxy-terminal domain RNA polymerase II polypeptide A small phosphatase 2 | | Descriptor: | Carboxy-terminal domain RNA polymerase II polypeptide A small phosphatase 2, MAGNESIUM ION | | Authors: | Bonanno, J.B, Dickey, M, Bain, K.T, Lau, C, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-05-31 | | Release date: | 2007-06-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structural genomics of protein phosphatases.
J.Struct.Funct.Genom., 8, 2007
|
|
9FMN
 
 | | Structure of Human PADI6 | | Descriptor: | Protein-arginine deiminase type-6 | | Authors: | Mouilleron, S, Walport, L, Williams, J, Marsh, A.J, Hernandez Trapero, R. | | Deposit date: | 2024-06-06 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Structural insight into the function of human peptidyl arginine deiminase 6.
Comput Struct Biotechnol J, 23, 2024
|
|
7AYY
 
 | | Structure of the human 8-oxoguanine DNA Glycosylase hOGG1 in complex with activator TH10785 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, N-glycosylase/DNA lyase, ... | | Authors: | Masuyer, G, Davies, J.R, Stenmark, P. | | Deposit date: | 2020-11-13 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Small-molecule activation of OGG1 increases oxidative DNA damage repair by gaining a new function.
Science, 376, 2022
|
|
7AYZ
 
 | |
7AZ0
 
 | |
6EHI
 
 | | NucT from Helicobacter pylori | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Celma, L, Li de la Sierra-Gallay, I, Quevillon-Cheruel, S. | | Deposit date: | 2017-09-13 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural basis for the substrate selectivity of Helicobacter pylori NucT nuclease activity.
PLoS ONE, 12, 2017
|
|
6ERG
 
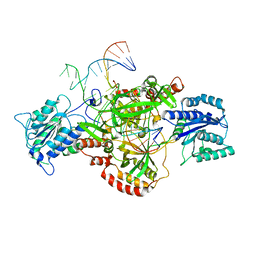 | | Complex of XLF and heterodimer Ku bound to DNA | | Descriptor: | DNA (21-MER), DNA (34-MER), Non-homologous end-joining factor 1, ... | | Authors: | Nemoz, C, Legrand, P, Ropars, V, Charbonnier, J.B. | | Deposit date: | 2017-10-18 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | XLF and APLF bind Ku80 at two remote sites to ensure DNA repair by non-homologous end joining.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6ERH
 
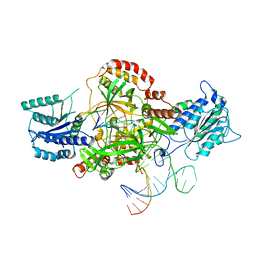 | | Complex of XLF and heterodimer Ku bound to DNA | | Descriptor: | DNA (21-MER), DNA (34-MER), Non-homologous end-joining factor 1, ... | | Authors: | Nemoz, C, Legrand, P, Ropars, V, Charbonnier, J.B. | | Deposit date: | 2017-10-18 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | XLF and APLF bind Ku80 at two remote sites to ensure DNA repair by non-homologous end joining.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6YEU
 
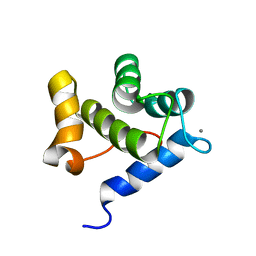 | | Second EH domain of AtEH1/Pan1 | | Descriptor: | CALCIUM ION, Calcium-binding EF hand family protein | | Authors: | Yperman, K, Papageorgiou, A, Evangelidis, T, Van Damme, D, Tripsianes, K. | | Deposit date: | 2020-03-25 | | Release date: | 2021-03-31 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Distinct EH domains of the endocytic TPLATE complex confer lipid and protein binding.
Nat Commun, 12, 2021
|
|
6YET
 
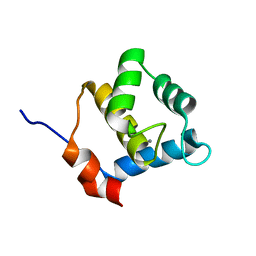 | | Second EH domain of AtEH1/Pan1 | | Descriptor: | CALCIUM ION, Calcium-binding EF hand family protein | | Authors: | Yperman, K, Papageorgiou, A, Evangelidis, T, Van Damme, D, Tripsianes, K. | | Deposit date: | 2020-03-25 | | Release date: | 2021-03-31 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Distinct EH domains of the endocytic TPLATE complex confer lipid and protein binding.
Nat Commun, 12, 2021
|
|
8P9Y
 
 | | SARS-CoV-2 S protein S:D614G mutant in 3-down with binding site of an entry inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SODIUM ION, ... | | Authors: | Adhav, A, Forcada-Nadal, A, Marco-Marin, C, Lopez-Redondo, M.L, Llacer, J.L. | | Deposit date: | 2023-06-06 | | Release date: | 2023-09-27 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | C-2 Thiophenyl Tryptophan Trimers Inhibit Cellular Entry of SARS-CoV-2 through Interaction with the Viral Spike (S) Protein.
J.Med.Chem., 66, 2023
|
|
8P99
 
 | | SARS-CoV-2 S-protein:D614G mutant in 1-up conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1,Spike glycoprotein | | Authors: | Adhav, A, Forcada-Nadal, A, Marco-Marin, C, Lopez-Redondo, M.L, Llacer, J.L. | | Deposit date: | 2023-06-05 | | Release date: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | C-2 Thiophenyl Tryptophan Trimers Inhibit Cellular Entry of SARS-CoV-2 through Interaction with the Viral Spike (S) Protein.
J.Med.Chem., 66, 2023
|
|
8PAR
 
 | | Crystal structure of human MAP4K1 with an inhibitor, BAY-405 | | Descriptor: | GLYCEROL, Mitogen-activated protein kinase kinase kinase kinase 1, ~{N}-[3,5-bis(fluoranyl)-4-[[3-[1-(trifluoromethyl)cyclopropyl]-1~{H}-pyrrolo[2,3-b]pyridin-4-yl]oxy]phenyl]-2,9-dioxa-4-azaspiro[5.5]undec-3-en-3-amine | | Authors: | Schaefer, M. | | Deposit date: | 2023-06-08 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification and optimization of Azaindole based MAP4K1 Inhibitors and the discovery of BAY-405
To Be Published
|
|
8PAS
 
 | | Crystal structure of MAP4K1 with a SMOL inhibitor | | Descriptor: | 4-[2,6-bis(fluoranyl)-4-(3-morpholin-4-ylpropylcarbamoylamino)phenoxy]-~{N}-[(4-methyl-1,2,5-oxadiazol-3-yl)methyl]-1~{H}-pyrrolo[2,3-b]pyridine-3-carboxamide, Mitogen-activated protein kinase kinase kinase kinase 1 | | Authors: | Friberg, A. | | Deposit date: | 2023-06-08 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Identification and optimization of Azaindole based MAP4K1 Inhibitors and the discovery of BAY-405
To Be Published
|
|
