5RYD
 
 | | INPP5D PanDDA analysis group deposition -- Crystal Structure of the phosphatase and C2 domains of SHIP1 in complex with Z955369596 | | Descriptor: | DIMETHYL SULFOXIDE, N-[(5-bromo-2-methoxyphenyl)methyl]acetamide, Phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 1 | | Authors: | Bradshaw, W.J, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Regulation of inositol 5-phosphatase activity by the C2 domain of SHIP1 and SHIP2.
Structure, 2024
|
|
5RYH
 
 | | INPP5D PanDDA analysis group deposition -- Crystal Structure of the phosphatase and C2 domains of SHIP1 in complex with Z1212984951 | | Descriptor: | (3S)-3-(methylamino)-1-phenylpiperidin-2-one, DIMETHYL SULFOXIDE, Phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 1 | | Authors: | Bradshaw, W.J, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Regulation of inositol 5-phosphatase activity by the C2 domain of SHIP1 and SHIP2.
Structure, 2024
|
|
5RL9
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 helicase in complex with Z1703168683 | | Descriptor: | 1-(3-fluoro-4-methylphenyl)methanesulfonamide, Helicase, PHOSPHATE ION, ... | | Authors: | Newman, J.A, Yosaatmadja, Y, Douangamath, A, Aimon, A, Powell, A.J, Dias, A, Fearon, D, Dunnett, L, Brandao-Neto, J, Krojer, T, Skyner, R, Gorrie-Stone, T, Thompson, W, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-09-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.788 Å) | | Cite: | Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase.
Nat Commun, 12, 2021
|
|
5RLS
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 helicase in complex with Z59181945 | | Descriptor: | Helicase, N-hydroxyquinoline-2-carboxamide, PHOSPHATE ION, ... | | Authors: | Newman, J.A, Yosaatmadja, Y, Douangamath, A, Aimon, A, Powell, A.J, Dias, A, Fearon, D, Dunnett, L, Brandao-Neto, J, Krojer, T, Skyner, R, Gorrie-Stone, T, Thompson, W, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-09-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.278 Å) | | Cite: | Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase.
Nat Commun, 12, 2021
|
|
5RMD
 
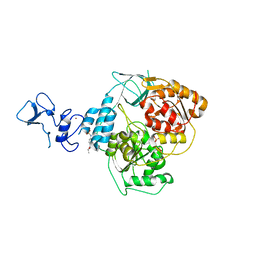 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 helicase in complex with Z57614330 | | Descriptor: | Helicase, N-ethyl-4-[(methylsulfonyl)amino]benzamide, PHOSPHATE ION, ... | | Authors: | Newman, J.A, Yosaatmadja, Y, Douangamath, A, Aimon, A, Powell, A.J, Dias, A, Fearon, D, Dunnett, L, Brandao-Neto, J, Krojer, T, Skyner, R, Gorrie-Stone, T, Thompson, W, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-09-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase.
Nat Commun, 12, 2021
|
|
5TB6
 
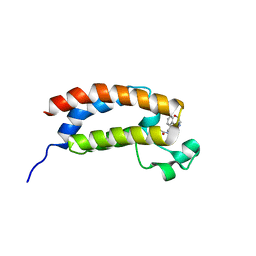 | | Structure of bromodomain of CREBBP with a pyrazolo[4,3-c]pyridin fragment | | Descriptor: | 1,2-ETHANEDIOL, 1-(3-phenyl-1,4,6,7-tetrahydropyrazolo[4,3-c]pyridin-5-yl)propan-1-one, CREB-binding protein | | Authors: | Filippakopoulos, P, Picaud, S, Knapp, S, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-09-11 | | Release date: | 2016-10-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Discovery of New Bromodomain Scaffolds by Biosensor Fragment Screening.
ACS Med Chem Lett, 7, 2016
|
|
5T5G
 
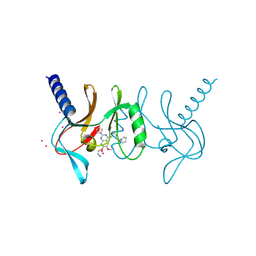 | | human SETD8 in complex with MS2177 | | Descriptor: | 7-(2-aminoethoxy)-6-methoxy-2-(pyrrolidin-1-yl)-N-[5-(pyrrolidin-1-yl)pentyl]quinazolin-4-amine, N-lysine methyltransferase KMT5A, UNKNOWN ATOM OR ION | | Authors: | Yu, W, Tempel, W, Babault, N, Ma, A, Butler, K.V, Jin, J, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-08-30 | | Release date: | 2016-09-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Based Design of a Covalent Inhibitor of the SET Domain-Containing Protein 8 (SETD8) Lysine Methyltransferase.
J. Med. Chem., 59, 2016
|
|
5TTG
 
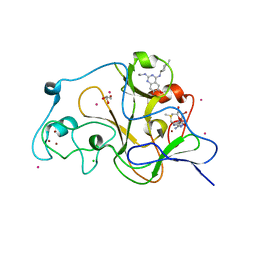 | | Crystal structure of catalytic domain of GLP with MS012 | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | DONG, A, ZENG, H, LIU, J, XIONG, Y, BABAULT, N, JIN, J, TEMPEL, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, WU, H, BROWN, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-11-03 | | Release date: | 2017-02-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Discovery of Potent and Selective Inhibitors for G9a-Like Protein (GLP) Lysine Methyltransferase.
J. Med. Chem., 60, 2017
|
|
5T1I
 
 | | CBX3 chromo shadow domain in complex with histone H3 peptide | | Descriptor: | Chromobox protein homolog 3, Histone H3.1, UNKNOWN ATOM OR ION | | Authors: | Liu, Y, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-08-19 | | Release date: | 2016-09-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Peptide recognition by heterochromatin protein 1 (HP1) chromoshadow domains revisited: Plasticity in the pseudosymmetric histone binding site of human HP1.
J. Biol. Chem., 292, 2017
|
|
5TTW
 
 | | Crystal Structure of EED in Complex with UNC4859 | | Descriptor: | Polycomb protein EED, SULFATE ION, UNC4859, ... | | Authors: | The, J, Barnash, K.D, Brown, P.J, Edwards, A.M, Bountra, C, Frye, S.V, James, L.I, Arrowsmith, C.H. | | Deposit date: | 2016-11-04 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Discovery of Peptidomimetic Ligands of EED as Allosteric Inhibitors of PRC2.
ACS Comb Sci, 19, 2017
|
|
5TPX
 
 | | Bromodomain from Plasmodium Faciparum Gcn5, complexed with compound | | Descriptor: | (1S,2S)-N~1~,N~1~-dimethyl-N~2~-(3-methyl[1,2,4]triazolo[3,4-a]phthalazin-6-yl)-1-phenylpropane-1,2-diamine, CHLORIDE ION, Histone acetyltransferase GCN5, ... | | Authors: | Lin, Y.H, Hou, C.F.D, MOUSTAKIM, M, DIXON, D.J, Loppnau, P, Tempel, W, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Hui, R, BRENNAN, P.E, Walker, J.R, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-10-21 | | Release date: | 2017-01-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of a PCAF Bromodomain Chemical Probe.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5TH7
 
 | | Complex of SETD8 with MS453 | | Descriptor: | 1,2-ETHANEDIOL, N-(3-{[6,7-dimethoxy-2-(pyrrolidin-1-yl)quinazolin-4-yl]amino}propyl)propanamide, N-lysine methyltransferase KMT5A, ... | | Authors: | Yu, W, Tempel, W, Babault, N, Ma, A, Butler, K.V, Jin, J, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-09-29 | | Release date: | 2016-11-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-Based Design of a Covalent Inhibitor of the SET Domain-Containing Protein 8 (SETD8) Lysine Methyltransferase.
J. Med. Chem., 59, 2016
|
|
5TTF
 
 | | Crystal structure of catalytic domain of G9a with MS012 | | Descriptor: | CHLORIDE ION, Histone-lysine N-methyltransferase EHMT2, N4-(1-methylpiperidin-4-yl)-N2-hexyl-6,7-dimethoxyquinazoline-2,4-diamine, ... | | Authors: | DONG, A, ZENG, H, LIU, J, XIONG, Y, BABAULT, N, JIN, J, TEMPEL, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, WU, H, BROWN, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-11-03 | | Release date: | 2016-12-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Discovery of Potent and Selective Inhibitors for G9a-Like Protein (GLP) Lysine Methyltransferase.
J. Med. Chem., 60, 2017
|
|
5O2D
 
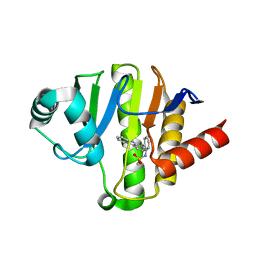 | | PARP14 Macrodomain 2 with inhibitor | | Descriptor: | Poly [ADP-ribose] polymerase 14, ~{N}-[2-(9~{H}-carbazol-1-yl)phenyl]methanesulfonamide | | Authors: | Uth, K, Schuller, M, Sieg, C, Wang, J, Krojer, T, Knapp, S, Riedels, K, Bracher, F, Edwards, A.M, Arrowsmith, C, Bountra, C, Elkins, J.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-05-20 | | Release date: | 2017-11-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of a Selective Allosteric Inhibitor Targeting Macrodomain 2 of Polyadenosine-Diphosphate-Ribose Polymerase 14.
ACS Chem. Biol., 12, 2017
|
|
5NFJ
 
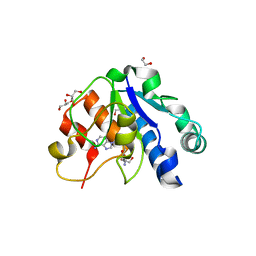 | | Crystal structure of the methyltransferase subunit of human mitochondrial Ribonuclease P (MRPP1) bound to S-adenosyl-methionine (SAM) | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Mitochondrial ribonuclease P protein 1, ... | | Authors: | Oerum, S, Kopec, J, Fitzpatrick, F, Newman, J.A, Chalk, R, Shrestha, L, Fairhead, M, Talon, R, Burgess-Brown, N, von Delft, F, Arrowsmith, C, Edwards, C, Bountra, C, Oppermann, U, Yue, W.W, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-03-14 | | Release date: | 2018-05-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal structure of the methyltransferase subunit of human mitochondrial Ribonuclease P (MRPP1) bound to S-adenosyl-methionine (SAM)
To Be Published
|
|
5O1P
 
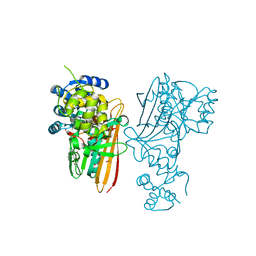 | | Crystal structure of human aminoadipate semialdehyde synthase, saccharopine dehydrogenase. | | Descriptor: | 1,2-ETHANEDIOL, Alpha-aminoadipic semialdehyde synthase, mitochondrial, ... | | Authors: | Kopec, J, Rembeza, E, Pena, I.A, Williams, E, Velupillai, S, Kupinska, K, Strain-Damerell, C, Goubin, S, Talon, R, Collins, P, Krojer, T, Burgess-Brown, N, Arrowsmith, C, Edwards, A, Bountra, C, von Delft, F, Arruda, P, Yue, W.W. | | Deposit date: | 2017-05-18 | | Release date: | 2017-06-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of human aminoadipate semialdehyde synthase, saccharopine dehydrogenase.
To Be Published
|
|
5O1O
 
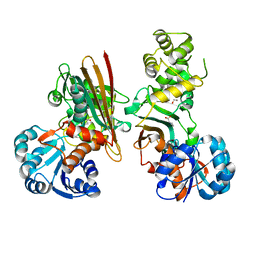 | | Crystal structure of human aminoadipate semialdehyde synthase, saccharopine dehydrogenase domain with proline bound. | | Descriptor: | 1,2-ETHANEDIOL, Alpha-aminoadipic semialdehyde synthase, mitochondrial, ... | | Authors: | Kopec, J, Rembeza, E, Pena, I.A, Mathea, S, Velupillai, S, Strain-Damerell, C, Goubin, S, Kupinska, K, Talon, R, Collins, P, Krojer, T, Burgess-Brown, N, Arrowsmith, C, Edwards, A, Bountra, C, von Delft, F, Arruda, P, Yue, W.W. | | Deposit date: | 2017-05-18 | | Release date: | 2017-06-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Crystal structure of human aminoadipate semialdehyde synthase, saccharopine dehydrogenase domain with proline bound.
To Be Published
|
|
5O1N
 
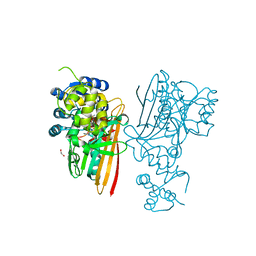 | | Crystal structure of human aminoadipate semialdehyde synthase, saccharopine dehydrogenase domain with N-[(2S)-2-Pyrrolidinylmethyl]-trifluoromethanesulfonamide bound | | Descriptor: | 1,1,1-tris(fluoranyl)-~{N}-[[(2~{S})-pyrrolidin-2-yl]methyl]methanesulfonamide, 1,2-ETHANEDIOL, Alpha-aminoadipic semialdehyde synthase, ... | | Authors: | Kopec, J, Rembeza, E, Pena, I.A, Strain-Damerell, C, Goubin, S, Sethi, R, Velupillai, S, Talon, R, Collins, P, Krojer, T, McLaughlin, M, Burgess-Brown, N, Arrowsmith, C, Edwards, A, Bountra, C, Brennan, P, von Delft, F, Arruda, P, Yue, W.W. | | Deposit date: | 2017-05-18 | | Release date: | 2017-05-31 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal structure of human aminoadipate semialdehyde synthase, saccharopine dehydrogenase domain with N-[(2S)-2-Pyrrolidinylmethyl]-trifluoromethanesulfonamide bound
To Be Published
|
|
5O5E
 
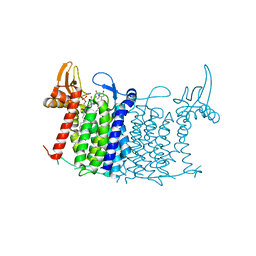 | | Crystal structure of human UDP-N-acetylglucosamine-dolichyl-phosphate N-acetylglucosaminephosphotransferase (DPAGT1) (V264G mutant) in complex with tunicamycin | | Descriptor: | (2S)-3-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-2-[(6E)-HEXADEC-6-ENOYLOXY]PROPYL (8E)-OCTADEC-8-ENOATE, Tunicamycin, UDP-N-acetylglucosamine--dolichyl-phosphate N-acetylglucosaminephosphotransferase, ... | | Authors: | Pike, A.C.W, Dong, Y.Y, Chu, A, Tessitore, A, Goubin, S, Dong, L, Mukhopadhyay, S, Mahajan, P, Chalk, R, Berridge, G, Wang, D, Kupinska, K, Belaya, K, Beeson, D, Burgess-Brown, N, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-06-01 | | Release date: | 2018-02-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structures of DPAGT1 Explain Glycosylation Disease Mechanisms and Advance TB Antibiotic Design.
Cell, 175, 2018
|
|
5OWR
 
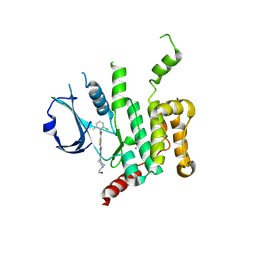 | | Human STK10 bound to dasatinib | | Descriptor: | CALCIUM ION, N-(2-CHLORO-6-METHYLPHENYL)-2-({6-[4-(2-HYDROXYETHYL)PIPERAZIN-1-YL]-2-METHYLPYRIMIDIN-4-YL}AMINO)-1,3-THIAZOLE-5-CARBOXAMIDE, Serine/threonine-protein kinase 10 | | Authors: | Szklarz, M, Muniz, J.R.C, Vollmar, M, von Delft, F, Bountra, C, Knapp, S, Edwards, A.M, Arrowsmith, C, Elkins, J.M. | | Deposit date: | 2017-09-04 | | Release date: | 2017-09-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Human STK10 bound to dasatinib
To Be Published
|
|
5OWQ
 
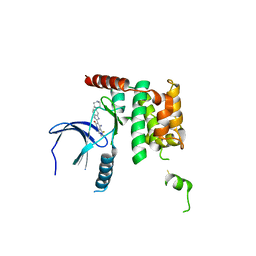 | | Human STK10 bound to dovitinib | | Descriptor: | 4-amino-5-fluoro-3-[5-(4-methylpiperazin-1-yl)-1H-benzimidazol-2-yl]quinolin-2(1H)-one, Serine/threonine-protein kinase 10 | | Authors: | Szklarz, M, von Delft, F, Bountra, C, Knapp, S, Edwards, A.M, Arrowsmith, C, Elkins, J.M. | | Deposit date: | 2017-09-04 | | Release date: | 2017-09-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Human STK10 bound to dovitinib
To Be Published
|
|
5PDO
 
 | | PanDDA analysis group deposition -- Crystal Structure of BAZ2B after initial refinement with no ligand modelled (structure 81) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-03 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5PE3
 
 | | PanDDA analysis group deposition -- Crystal Structure of BAZ2B after initial refinement with no ligand modelled (structure 96) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-03 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5PEN
 
 | | PanDDA analysis group deposition -- Crystal Structure of BAZ2B after initial refinement with no ligand modelled (structure 116) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-03 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5PBS
 
 | | PanDDA analysis group deposition -- Crystal Structure of BAZ2B after initial refinement with no ligand modelled (structure 13) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-03 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
