5K5F
 
 | | NMR structure of the HLTF HIRAN domain | | Descriptor: | Helicase-like transcription factor | | Authors: | Bezsonova, I, Neculai, D, Korzhnev, D, Weigelt, J, Bountra, C, Edwards, A, Arrowsmith, C, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-05-23 | | Release date: | 2016-06-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the HLTF HIRAN domain
To Be Published
|
|
1MZ4
 
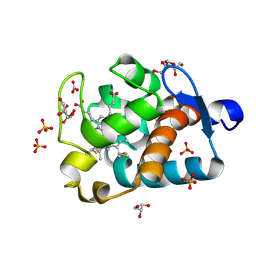 | | Crystal Structure of Cytochrome c550 from Thermosynechococcus elongatus | | Descriptor: | BICARBONATE ION, GLYCEROL, HEME C, ... | | Authors: | Kerfeld, C.A, Sawaya, M.R, Bottin, H, Tran, K.T, Sugiura, M, Kirilovsky, D, Krogmann, D, Yeates, T.O, Boussac, A. | | Deposit date: | 2002-10-05 | | Release date: | 2003-09-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and EPR characterization of the soluble form of cytochrome c-550 and of the psbV2 gene product from the cyanobacterium Thermosynechococcus elongatus.
Plant Cell.Physiol., 44, 2003
|
|
6RQP
 
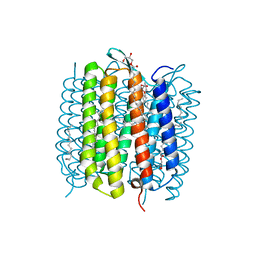 | | Steady-state-SMX dark state structure of bacteriorhodopsin | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, Bacteriorhodopsin, ... | | Authors: | Weinert, T, Skopintsev, P, James, D, Kekilli, D, Furrer, A, Bruenle, S, Mous, S, Nogly, P, Standfuss, J. | | Deposit date: | 2019-05-16 | | Release date: | 2019-07-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Proton uptake mechanism in bacteriorhodopsin captured by serial synchrotron crystallography.
Science, 365, 2019
|
|
1RNQ
 
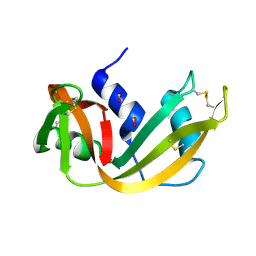 | | RIBONUCLEASE A CRYSTALLIZED FROM 8M SODIUM FORMATE | | Descriptor: | FORMIC ACID, RIBONUCLEASE A | | Authors: | Fedorov, A.A, Josef-Mccarthy, D, Graf, I, Anguelova, D, Fedorov, E.V, Almo, S.C. | | Deposit date: | 1995-11-08 | | Release date: | 1996-04-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ionic interactions in crystalline bovine pancreatic ribonuclease A.
Biochemistry, 35, 1996
|
|
1HFD
 
 | | HUMAN COMPLEMENT FACTOR D IN A P21 CRYSTAL FORM | | Descriptor: | COMPLEMENT FACTOR D | | Authors: | Jing, H, Babu, Y.S, Moore, D, Kilpatrick, J.M, Liu, X.-Y, Volanakis, J.E, Narayana, S.V.L. | | Deposit date: | 1998-06-18 | | Release date: | 1999-06-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of native and complexed complement factor D: implications of the atypical His57 conformation and self-inhibitory loop in the regulation of specific serine protease activity.
J.Mol.Biol., 282, 1998
|
|
7O4W
 
 | |
9FH4
 
 | | Cryo-EM Structure of Amyloid-beta Fibrils Carrying the Uppsala AbetaUpp(1-42)delta(19-24) Mutation - Polymorph 3 | | Descriptor: | Amyloid-beta precursor protein | | Authors: | Zielinski, M, Peralta Reyes, F.S, Gremer, L, Pagnon de la Vega, M, Roeder, C, Heidler, T.V, Syvaenen, S, Willbold, D, Sehlin, D, Ingelsson, M, Schroeder, G.F. | | Deposit date: | 2024-05-26 | | Release date: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM Structures of Amyloid-beta Fibrils from human and murine brains carrying the Uppsala AbetaUpp(1-42)delta(19-24) mutation
To Be Published
|
|
9FH5
 
 | | Cryo-EM Structure of Amyloid-beta Fibrils Carrying the Uppsala AbetaUpp(1-42)delta(19-24) Mutation - Polymorph 4 | | Descriptor: | Amyloid-beta precursor protein | | Authors: | Zielinski, M, Peralta Reyes, F.S, Gremer, L, Pagnon de la Vega, M, Roeder, C, Heidler, T.V, Syvaenen, S, Willbold, D, Sehlin, D, Ingelsson, M, Schroeder, G.F. | | Deposit date: | 2024-05-26 | | Release date: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM Structures of Amyloid-beta Fibrils from human and murine brains carrying the Uppsala AbetaUpp(1-42)delta(19-24) mutation
To Be Published
|
|
9FH2
 
 | | Cryo-EM Structure of Amyloid-beta Fibrils Carrying the Uppsala AbetaUpp(1-42)delta(19-24) Mutation - Polymorph 1 | | Descriptor: | Amyloid-beta precursor protein | | Authors: | Zielinski, M, Peralta Reyes, F.S, Gremer, L, Pagnon de la Vega, M, Roeder, C, Heidler, T.V, Syvaenen, S, Willbold, D, Sehlin, D, Ingelsson, M, Schroeder, G.F. | | Deposit date: | 2024-05-26 | | Release date: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM Structures of Amyloid-beta Fibrils from human and murine brains carrying the Uppsala AbetaUpp(1-42)delta(19-24) mutation
To Be Published
|
|
9GD8
 
 | |
9FH1
 
 | | Cryo-EM Structure of Amyloid-beta Fibrils from Mouse Brain Carrying the Uppsala AbetaUpp(1-42)delta(19-24) Mutation | | Descriptor: | Amyloid-beta precursor protein | | Authors: | Zielinski, M, Peralta Reyes, F.S, Gremer, L, Pagnon de la Vega, M, Roeder, C, Heidler, T.V, Syvaenen, S, Willbold, D, Sehlin, D, Ingelsson, M, Schroeder, G.F. | | Deposit date: | 2024-05-26 | | Release date: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM Structures of Amyloid-beta Fibrils from human and murine brains carrying the Uppsala AbetaUpp(1-42)delta(19-24) mutation
To Be Published
|
|
8PT4
 
 | | beta-Ureidopropionase tetramer | | Descriptor: | Beta-ureidopropionase | | Authors: | Cederfelt, D, Dobritzsch, D. | | Deposit date: | 2023-07-13 | | Release date: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | The Allosteric Regulation of Beta-Ureidopropionase Depends on Fine-Tuned Stability of Active-Site Loops and Subunit Interfaces.
Biomolecules, 13, 2023
|
|
9FQW
 
 | | Cryo-EM structure of MmCAT1 bound with FrMLV-RBD in the ornithine-bound inward-occluded state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, High affinity cationic amino acid transporter 1,Green fluorescent protein, ... | | Authors: | Ye, M, Zhou, D, Pike, A.C.W, Wang, S, Wang, D, Bakshi, S, Brooke, L, Williams, E, Elkins, J, Stuart, D.I, Sauer, D.B. | | Deposit date: | 2024-06-17 | | Release date: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Amino acid and viral binding by the high-affinity Cationic Amino acid Transporter 1 (CAT1) from Mus musculus
To Be Published
|
|
6RPH
 
 | | TR-SMX open state structure (10-15ms) of bacteriorhodopsin | | Descriptor: | Bacteriorhodopsin, RETINAL | | Authors: | Weinert, T, Skopintsev, P, James, D, Kekilli, D, Furrer, A, Bruenle, S, Mous, S, Nogly, P, Standfuss, J. | | Deposit date: | 2019-05-14 | | Release date: | 2019-07-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Proton uptake mechanism in bacteriorhodopsin captured by serial synchrotron crystallography.
Science, 365, 2019
|
|
9FH6
 
 | | Cryo-EM Structure of Tau Filaments from Individuals Carrying the Uppsala AbetaUpp(1-42)delta(19-24) Mutation | | Descriptor: | Microtubule-associated protein tau | | Authors: | Zielinski, M, Peralta Reyes, F.S, Gremer, L, Pagnon de la Vega, M, Roeder, C, Heidler, T.V, Syvaenen, S, Willbold, D, Sehlin, D, Ingelsson, M, Schroeder, G.F. | | Deposit date: | 2024-05-26 | | Release date: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM Structures of Amyloid-beta Fibrils from human and murine brains carrying the Uppsala AbetaUpp(1-42)delta(19-24) mutation
To Be Published
|
|
9GD9
 
 | |
9GD6
 
 | |
1DTP
 
 | |
9FH3
 
 | | Cryo-EM Structure of Amyloid-beta Fibrils Carrying the Uppsala AbetaUpp(1-42)delta(19-24) Mutation - Polymorph 2 | | Descriptor: | Amyloid-beta precursor protein | | Authors: | Zielinski, M, Peralta Reyes, F.S, Gremer, L, Pagnon de la Vega, M, Roeder, C, Heidler, T.V, Syvaenen, S, Willbold, D, Sehlin, D, Ingelsson, M, Schroeder, G.F. | | Deposit date: | 2024-05-26 | | Release date: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM Structures of Amyloid-beta Fibrils from human and murine brains carrying the Uppsala AbetaUpp(1-42)delta(19-24) mutation
To Be Published
|
|
9FQU
 
 | | Cryo-EM structure of MmCAT1 bound with FrMLV-RBD in the arginine-bound inward-occluded state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ARGININE, ... | | Authors: | Ye, M, Zhou, D, Pike, A.C.W, Wang, S, Wang, D, Bakshi, S, Brooke, L, Williams, E, Elkins, J, Stuart, D.I, Sauer, D.B. | | Deposit date: | 2024-06-17 | | Release date: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Amino acid and viral binding by the high-affinity Cationic Amino acid Transporter 1 (CAT1) from Mus musculus
To Be Published
|
|
5HMS
 
 | | X-ray structure of human recombinant 5-aminolaevulinic acid dehydratase (hrALAD). | | Descriptor: | Delta-aminolevulinic acid dehydratase, ZINC ION | | Authors: | Butler, D, Erskine, P.T, Cooper, J.B, Shoolingin-Jordan, P.M. | | Deposit date: | 2016-01-17 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural studies of substrate and product complexes of 5-aminolaevulinic acid dehydratase from humans, Escherichia coli and the hyperthermophile Pyrobaculum calidifontis.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
9FQT
 
 | | Cryo-EM structure of MmCAT1 bound with FrMLV-RBD in the apo inward-open state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Ye, M, Zhou, D, Pike, A.C.W, Wang, S, Wang, D, Bakshi, S, Brooke, L, Williams, E, Elkins, J, Stuart, D.I, Sauer, D.B. | | Deposit date: | 2024-06-17 | | Release date: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Amino acid and viral binding by the high-affinity Cationic Amino acid Transporter 1 (CAT1) from Mus musculus
To Be Published
|
|
9FQV
 
 | | Cryo-EM structure of MmCAT1 bound with FrMLV-RBD in the lysine-bound inward-occluded state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, High affinity cationic amino acid transporter 1,Green fluorescent protein, ... | | Authors: | Ye, M, Zhou, D, Pike, A.C.W, Wang, S, Wang, D, Bakshi, S, Brooke, L, Williams, E, Elkins, J, Stuart, D.I, Sauer, D.B. | | Deposit date: | 2024-06-17 | | Release date: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Amino acid and viral binding by the high-affinity Cationic Amino acid Transporter 1 (CAT1) from Mus musculus
To Be Published
|
|
5VWR
 
 | | E.coli Aspartate aminotransferase-(1R,3S,4S)-3-amino-4-fluorocyclopentane-1-carboxylic acid (FCP)-alpha-ketoglutarate | | Descriptor: | (E)-N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-L-glutamic acid, Aspartate aminotransferase, GLYCEROL | | Authors: | Mascarenhas, R, Liu, D, Le, H, Silverman, R. | | Deposit date: | 2017-05-22 | | Release date: | 2017-09-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Selective Targeting by a Mechanism-Based Inactivator against Pyridoxal 5'-Phosphate-Dependent Enzymes: Mechanisms of Inactivation and Alternative Turnover.
Biochemistry, 56, 2017
|
|
5VZ4
 
 | | Receptor-growth factor crystal structure at 2.20 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, GDNF family receptor alpha-like, ... | | Authors: | Lakshminarasimhan, D, White, A, Suto, R.K. | | Deposit date: | 2017-05-26 | | Release date: | 2017-09-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Non-homeostatic body weight regulation through a brainstem-restricted receptor for GDF15.
Nature, 550, 2017
|
|
