6PDQ
 
 | | Ancestral Effector Caspase 3/6/7 | | Descriptor: | Ac-DEVD inhibitor, Ancestral Effector Caspase-3/6/7 | | Authors: | Clark, A.C. | | Deposit date: | 2019-06-19 | | Release date: | 2019-11-13 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Resurrection of ancestral effector caspases identifies novel networks for evolution of substrate specificity.
Biochem.J., 476, 2019
|
|
2P2I
 
 | | Crystal structure of the VEGFR2 kinase domain in complex with a nicotinamide inhibitor | | Descriptor: | N-(4-phenoxyphenyl)-2-[(pyridin-4-ylmethyl)amino]nicotinamide, Vascular endothelial growth factor receptor 2 | | Authors: | Whittington, D.A, Kim, J.L, Long, A.M, Rose, P, Gu, Y, Zhao, H. | | Deposit date: | 2007-03-07 | | Release date: | 2007-03-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Evolution of a Highly Selective and Potent 2-(Pyridin-2-yl)-1,3,5-triazine Tie-2 Kinase Inhibitor
J.Med.Chem., 50, 2007
|
|
6NQB
 
 | |
3IBE
 
 | | Crystal Structure of a Pyrazolopyrimidine Inhibitor Bound to PI3 Kinase Gamma | | Descriptor: | 1-(4-{4-morpholin-4-yl-1-[1-(pyridin-3-ylcarbonyl)piperidin-4-yl]-1H-pyrazolo[3,4-d]pyrimidin-6-yl}phenyl)-3-pyridin-4-ylurea, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Bard, J, Svenson, K. | | Deposit date: | 2009-07-15 | | Release date: | 2009-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.798 Å) | | Cite: | ATP-Competitive Inhibitors of the Mammalian Target of Rapamycin: Design and Synthesis of Highly Potent and Selective Pyrazolopyrimidines.
J.Med.Chem., 52, 2009
|
|
6BBU
 
 | | Crystal Structure of JAK1 in complex with compound 25 | | Descriptor: | N-{cis-3-[methyl(7H-pyrrolo[2,3-d]pyrimidin-4-yl)amino]cyclobutyl}propane-1-sulfonamide, Tyrosine-protein kinase JAK1 | | Authors: | Han, S. | | Deposit date: | 2017-10-19 | | Release date: | 2018-01-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Identification of N-{cis-3-[Methyl(7H-pyrrolo[2,3-d]pyrimidin-4-yl)amino]cyclobutyl}propane-1-sulfonamide (PF-04965842): A Selective JAK1 Clinical Candidate for the Treatment of Autoimmune Diseases.
J. Med. Chem., 61, 2018
|
|
2OO8
 
 | | Synthesis, Structural Analysis, and SAR Studies of Triazine Derivatives as Potent, Selective Tie-2 Inhibitors | | Descriptor: | Angiopoietin-1 receptor, N-{3-[3-(DIMETHYLAMINO)PROPYL]-5-(TRIFLUOROMETHYL)PHENYL}-4-METHYL-3-[(3-PYRIMIDIN-4-YLPYRIDIN-2-YL)AMINO]BENZAMIDE | | Authors: | Bellon, S.F, Kim, J. | | Deposit date: | 2007-01-25 | | Release date: | 2007-03-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Synthesis, structural analysis, and SAR studies of triazine derivatives as potent, selective Tie-2 inhibitors.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
6X3N
 
 | | Co-structure of BTK kinase domain with L-005085737 inhibitor | | Descriptor: | 4-[8-azanyl-3-[(2~{S})-1-[4-(dimethylamino)butanoyl]pyrrolidin-2-yl]imidazo[1,5-a]pyrazin-1-yl]-~{N}-(1,3-thiazol-2-yl)benzamide, 4-{8-amino-3-[(6R,8aS)-3-oxohexahydro-3H-[1,3]oxazolo[3,4-a]pyridin-6-yl]imidazo[1,5-a]pyrazin-1-yl}-N-[4-(trifluoromethyl)pyridin-2-yl]benzamide, Tyrosine-protein kinase BTK | | Authors: | Fischmann, T.O. | | Deposit date: | 2020-05-21 | | Release date: | 2020-07-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Potent, non-covalent reversible BTK inhibitors with 8-amino-imidazo[1,5-a]pyrazine core featuring 3-position bicyclic ring substitutes.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
6ZH0
 
 | | Structure of human galactokinase 1 bound with 2-(4-chlorophenyl)-N-(pyrimidin-2-yl)acetamide | | Descriptor: | 2-(1,3-benzoxazol-2-ylamino)spiro[1,6,7,8-tetrahydroquinazoline-4,1'-cyclohexane]-5-one, Galactokinase, N-(3-chlorophenyl)-2,2,2-trifluoroacetamide, ... | | Authors: | Mackinnon, S.R, Bezerra, G.A, Zhang, M, Foster, W, Krojer, T, Brandao-Neto, J, Douangamath, A, Arrowsmith, C, Edwards, A, Bountra, C, Brennan, P, Lai, K, Yue, W.W. | | Deposit date: | 2020-06-20 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Fragment Screening Reveals Starting Points for Rational Design of Galactokinase 1 Inhibitors to Treat Classic Galactosemia.
Acs Chem.Biol., 16, 2021
|
|
6X3O
 
 | | Co-structure of BTK kinase domain with L-005191930 inhibitor | | Descriptor: | 4-[8-azanyl-3-[(2~{S})-1-[4-(dimethylamino)butanoyl]pyrrolidin-2-yl]imidazo[1,5-a]pyrazin-1-yl]-~{N}-(1,3-thiazol-2-yl)benzamide, 4-{8-amino-3-[(6R,8aS)-3-oxo-3,5,6,7,8,8a-hexahydroindolizin-6-yl]imidazo[1,5-a]pyrazin-1-yl}-3-methoxy-N-[4-(trifluoromethyl)pyridin-2-yl]benzamide, Tyrosine-protein kinase BTK | | Authors: | Fischmann, T.O. | | Deposit date: | 2020-05-21 | | Release date: | 2020-07-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Potent, non-covalent reversible BTK inhibitors with 8-amino-imidazo[1,5-a]pyrazine core featuring 3-position bicyclic ring substitutes.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
8F1G
 
 | | Crystal structure of human WDR5 in complex with compound WM662 | | Descriptor: | (2S)-2-({(2S)-3-(3'-chloro[1,1'-biphenyl]-4-yl)-1-oxo-1-[(1H-tetrazol-5-yl)amino]propan-2-yl}oxy)propanoic acid, GLYCEROL, SULFATE ION, ... | | Authors: | Liu, H. | | Deposit date: | 2022-11-05 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Discovery of Potent Small-Molecule Inhibitors of WDR5-MYC Interaction.
Acs Chem.Biol., 18, 2023
|
|
7OO6
 
 | | Mechanosensitive channel MscS solubilized with DDM in closed conformation with added lipid | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, DODECYL-BETA-D-MALTOSIDE, Mechanosensitive channel of small conductance (MscS) | | Authors: | Rasmussen, T, Flegler, V.J, Boettcher, B. | | Deposit date: | 2021-05-26 | | Release date: | 2021-09-01 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Mechanosensitive channel gating by delipidation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7OOA
 
 | | Mechanosensitive channel MscS solubilized with LMNG in open conformation with added lipid | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, DODECYL-BETA-D-MALTOSIDE, Lauryl Maltose Neopentyl Glycol, ... | | Authors: | Rasmussen, T, Flegler, V.J, Boettcher, B. | | Deposit date: | 2021-05-27 | | Release date: | 2021-09-01 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Mechanosensitive channel gating by delipidation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7OO0
 
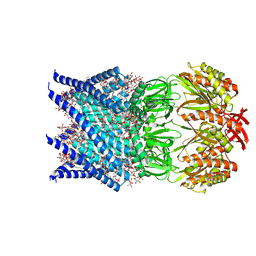 | | Mechanosensitive channel MscS solubilized with DDM in open conformation | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, DODECYL-BETA-D-MALTOSIDE, Mechanosensitive channel MscS | | Authors: | Rasmussen, T, Flegler, V.J, Boettcher, B. | | Deposit date: | 2021-05-26 | | Release date: | 2021-09-01 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Mechanosensitive channel gating by delipidation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7OO8
 
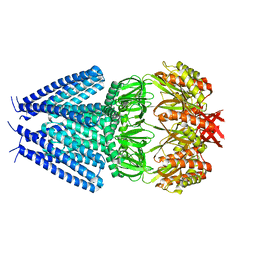 | |
7ONJ
 
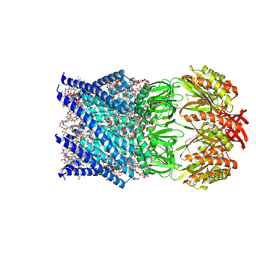 | | Mechanosensitive channel MscS solubilized with LMNG in open conformation | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, DODECYL-BETA-D-MALTOSIDE, Lauryl Maltose Neopentyl Glycol, ... | | Authors: | Rasmussen, T, Flegler, V.J, Boettcher, B. | | Deposit date: | 2021-05-25 | | Release date: | 2021-09-01 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Mechanosensitive channel gating by delipidation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7ONL
 
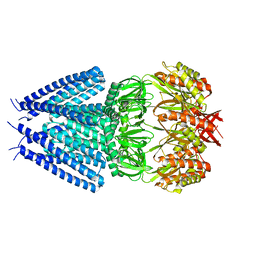 | |
6YS3
 
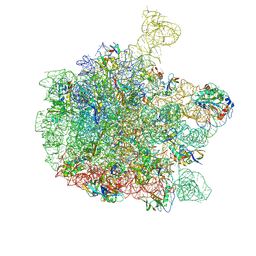 | | Cryo-EM structure of the 50S ribosomal subunit at 2.58 Angstroms with modeled GBC SecM peptide | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Schulte, L, Reitz, J, Kudlinzki, D, Hodirnau, V.V, Frangakis, A, Schwalbe, H. | | Deposit date: | 2020-04-20 | | Release date: | 2020-09-30 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.58 Å) | | Cite: | Cryo-EM structure of the 50S ribosomal subunit at 2.58 Angstroms with modeled GBC SecM peptide
Nat Commun, 2020
|
|
4RLO
 
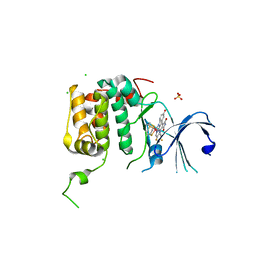 | | Human p70s6k1 with ruthenium-based inhibitor EM5 | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Domsic, J.F, Barber-Rotenberg, J, Salami, J, Qin, J, Marmorstein, R. | | Deposit date: | 2014-10-17 | | Release date: | 2015-01-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.527 Å) | | Cite: | Development of Organometallic S6K1 Inhibitors.
J.Med.Chem., 58, 2015
|
|
8PH4
 
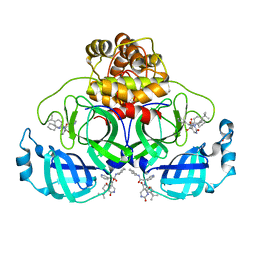 | | Co-Crystal structure of the SARS-CoV2 main protease Nsp5 with an Uracil-carrying X77-like inhibitor | | Descriptor: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, MALONATE ION, ... | | Authors: | Barthel, T, Altincekic, N, Jores, N, Wollenhaupt, J, Weiss, M.S, Schwalbe, H. | | Deposit date: | 2023-06-18 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Targeting the Main Protease (M pro , nsp5) by Growth of Fragment Scaffolds Exploiting Structure-Based Methodologies.
Acs Chem.Biol., 19, 2024
|
|
8PFP
 
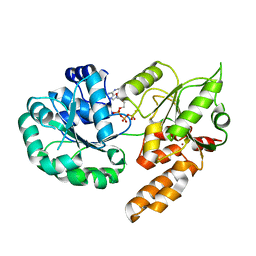 | |
8PFL
 
 | | Crystal structure of WRN helicase domain in complex with 3 | | Descriptor: | 2-[2-(3,6-dihydro-2~{H}-pyran-4-yl)-5-ethyl-7-oxidanylidene-6-[4-(3-oxidanylpyridin-2-yl)carbonylpiperazin-1-yl]-[1,2,4]triazolo[1,5-a]pyrimidin-4-yl]-~{N}-[2-methyl-4-(trifluoromethyl)phenyl]ethanamide, Bifunctional 3'-5' exonuclease/ATP-dependent helicase WRN, ZINC ION | | Authors: | Scheufler, C, Meyer, M, Moebitz, H. | | Deposit date: | 2023-06-16 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of WRN inhibitor HRO761 with synthetic lethality in MSI cancers.
Nature, 629, 2024
|
|
7MCS
 
 | | Cryo-electron microscopy structure of TnsC(1-503)A225V bound to DNA | | Descriptor: | DNA (5'-D(P*CP*GP*CP*GP*CP*GP*CP*GP*CP*GP*CP*GP*CP*GP*C)-3'), DNA (5'-D(P*GP*CP*GP*CP*GP*CP*GP*CP*GP*CP*GP*CP*GP*CP*G)-3'), MAGNESIUM ION, ... | | Authors: | Shen, Y, Ortega, J, Guarne, A. | | Deposit date: | 2021-04-02 | | Release date: | 2022-02-23 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | Structural basis for DNA targeting by the Tn7 transposon.
Nat.Struct.Mol.Biol., 29, 2022
|
|
8PFO
 
 | | Crystal structure of WRN helicase domain in complex with HRO761 | | Descriptor: | Bifunctional 3'-5' exonuclease/ATP-dependent helicase WRN, ZINC ION, ~{N}-[2-chloranyl-4-(trifluoromethyl)phenyl]-2-[2-(3,6-dihydro-2~{H}-pyran-4-yl)-5-ethyl-6-[4-(6-methyl-5-oxidanyl-pyrimidin-4-yl)carbonylpiperazin-1-yl]-7-oxidanylidene-[1,2,4]triazolo[1,5-a]pyrimidin-4-yl]ethanamide | | Authors: | Scheufler, C, Meyer, M, Moebitz, H. | | Deposit date: | 2023-06-16 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of WRN inhibitor HRO761 with synthetic lethality in MSI cancers.
Nature, 629, 2024
|
|
4RLP
 
 | | Human p70s6k1 with ruthenium-based inhibitor FL772 | | Descriptor: | CHLORIDE ION, [(amino-kappaN)methanethiolato](3-fluoro-9-methoxypyrido[2,3-a]pyrrolo[3,4-c]carbazole-5,7(6H,12H)-dionato-kappa~2~N,N')(N-methyl-1,4,7-trithiecan-9-amine-kappa~3~S~1~,S~4~,S~7~)ruthenium, p70S6K1 | | Authors: | Domsic, J.F, Barber-Rotenberg, J, Salami, J, Qin, J, Marmorstein, R. | | Deposit date: | 2014-10-17 | | Release date: | 2015-01-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Development of Organometallic S6K1 Inhibitors.
J.Med.Chem., 58, 2015
|
|
3U4E
 
 | | Crystal Structure of PG9 Fab in Complex with V1V2 Region from HIV-1 strain CAP45 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PG9 Heavy Chain, PG9 Light Chain, ... | | Authors: | Gorman, J, McLellan, J, Pancera, M, Kwong, P.D. | | Deposit date: | 2011-10-07 | | Release date: | 2011-11-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.185 Å) | | Cite: | Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9.
Nature, 480, 2011
|
|
