6QJ6
 
 | | The structure of Trehalose-6-phosphatase from Burkholderia pseudomallei | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Trehalose 6-phosphate phosphatase | | Authors: | Gourlay, L.J. | | Deposit date: | 2019-01-23 | | Release date: | 2020-01-15 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Functional and structural analysis of trehalose-6-phosphate phosphatase from Burkholderia pseudomallei: Insights into the catalytic mechanism.
Biochem.Biophys.Res.Commun., 523, 2020
|
|
8OUP
 
 | |
8CPE
 
 | |
1AOZ
 
 | | REFINED CRYSTAL STRUCTURE OF ASCORBATE OXIDASE AT 1.9 ANGSTROMS RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ASCORBATE OXIDASE, COPPER (II) ION, ... | | Authors: | Messerschmidt, A, Ladenstein, R, Huber, R. | | Deposit date: | 1992-01-08 | | Release date: | 1993-10-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Refined crystal structure of ascorbate oxidase at 1.9 A resolution.
J.Mol.Biol., 224, 1992
|
|
7PQ9
 
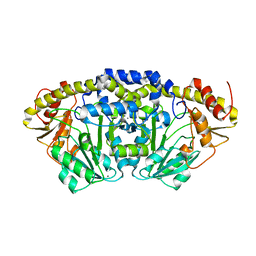 | | Crystal structure of Bacillus clausii pdxR at 2.8 Angstroms resolution | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Vivoli Vega, M, Isupov, M.N, Harmer, N. | | Deposit date: | 2021-09-16 | | Release date: | 2022-09-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into the DNA recognition mechanism by the bacterial transcription factor PdxR.
Nucleic Acids Res., 51, 2023
|
|
4OXC
 
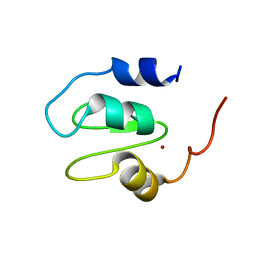 | | Crystal structure of XIAP BIR1 domain | | Descriptor: | E3 ubiquitin-protein ligase XIAP, ZINC ION | | Authors: | Milani, M, Cossu, F, Mastrangelo, E. | | Deposit date: | 2014-02-05 | | Release date: | 2015-02-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | NF023 binding to XIAP-BIR1: Searching drugs for regulation of the NF-kappa B pathway.
Proteins, 83, 2015
|
|
1PKY
 
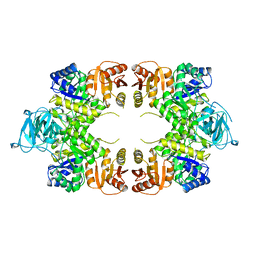 | | PYRUVATE KINASE FROM E. COLI IN THE T-STATE | | Descriptor: | PYRUVATE KINASE | | Authors: | Mattevi, A. | | Deposit date: | 1995-04-27 | | Release date: | 1995-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Escherichia coli pyruvate kinase type I: molecular basis of the allosteric transition.
Structure, 3, 1995
|
|
6G49
 
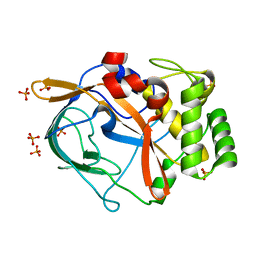 | | Crystal structure of the periplasmic domain of TgpA from Pseudomonas aeruginosa | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, Protein-glutamine gamma-glutamyltransferase | | Authors: | Milani, M, Mastrangelo, E, Uruburu, M. | | Deposit date: | 2018-03-27 | | Release date: | 2019-04-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and functional characterization of TgpA, a critical protein for the viability of Pseudomonas aeruginosa.
J.Struct.Biol., 205, 2019
|
|
6G4H
 
 | | Crystal structure of the periplasmic domain of TgpA from Pseudomonas aeruginosa bound to ethylmercury | | Descriptor: | ETHYL MERCURY ION, PHOSPHATE ION, Protein-glutamine gamma-glutamyltransferase | | Authors: | Milani, M, Mastrangelo, E, Uruburu, M. | | Deposit date: | 2018-03-27 | | Release date: | 2019-04-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and functional characterization of TgpA, a critical protein for the viability of Pseudomonas aeruginosa.
J.Struct.Biol., 205, 2019
|
|
9FAA
 
 | | Cryo-EM structure of cardiac collagen-associated amyloid AL59 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Monoclonal immunoglobulin light chains (LC) | | Authors: | Schulte, T, Speranzini, V, Chaves-Sanjuan, A, Milazzo, M, Ricagno, S. | | Deposit date: | 2024-05-10 | | Release date: | 2024-07-17 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Helical superstructures between amyloid and collagen in cardiac fibrils from a patient with AL amyloidosis.
Nat Commun, 15, 2024
|
|
9FAB
 
 | | Additional cryo-EM structure of cardiac amyloid AL59 - bent polymorph | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Monoclonal immunoglobulin light chains (LC) | | Authors: | Schulte, T, Speranzini, V, Chaves-Sanjuan, A, Milazzo, M, Ricagno, S. | | Deposit date: | 2024-05-10 | | Release date: | 2024-08-14 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Additional cryo-EM structure of cardiac amyloid AL59 - bent polymorph
To be published
|
|
9FAC
 
 | | Additional cryo-EM structure of cardiac amyloid AL59 - mixed polymorph | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Monoclonal immunoglobulin light chains (LC) | | Authors: | Schulte, T, Speranzini, V, Chaves-Sanjuan, A, Milazzo, M, Ricagno, S. | | Deposit date: | 2024-05-10 | | Release date: | 2024-08-14 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Helical superstructures between amyloid and collagen in cardiac fibrils from a patient with AL amyloidosis.
Nat Commun, 15, 2024
|
|
5M3T
 
 | | Structural tuning of CD81LEL (space group P64) | | Descriptor: | 1,2-ETHANEDIOL, CD81 antigen, CHLORIDE ION | | Authors: | Cunha, E.S, Sfriso, P, Rojas, A.L, Roversi, P, Hospital, A, Orozco, M, Abrescia, N.G. | | Deposit date: | 2016-10-17 | | Release date: | 2016-12-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.021 Å) | | Cite: | Mechanism of Structural Tuning of the Hepatitis C Virus Human Cellular Receptor CD81 Large Extracellular Loop.
Structure, 25, 2017
|
|
5M3D
 
 | | Structural tuning of CD81LEL (space group P31) | | Descriptor: | 1,2-ETHANEDIOL, CD81 antigen, PHOSPHATE ION | | Authors: | Cunha, E.S, Sfriso, P, Rojas, A.L, Roversi, P, Hospital, A, Orozco, M, Abrescia, N.G. | | Deposit date: | 2016-10-14 | | Release date: | 2016-12-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Mechanism of Structural Tuning of the Hepatitis C Virus Human Cellular Receptor CD81 Large Extracellular Loop.
Structure, 25, 2017
|
|
5M4R
 
 | | Structural tuning of CD81LEL (space group C2) | | Descriptor: | 1,2-ETHANEDIOL, CD81 antigen, SULFATE ION | | Authors: | Cunha, E.S, Sfriso, P, Rojas, A.L, Roversi, P, Hospital, A, Orozco, M, Abrescia, N.G. | | Deposit date: | 2016-10-19 | | Release date: | 2016-12-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Mechanism of Structural Tuning of the Hepatitis C Virus Human Cellular Receptor CD81 Large Extracellular Loop.
Structure, 25, 2017
|
|
5IQ6
 
 | | Crystal structure of Dengue virus serotype 3 RNA dependent RNA polymerase bound to HeE1-2Tyr, a new pyridobenzothizole inhibitor | | Descriptor: | N-[8-(cyclohexyloxy)-1-oxo-2-phenyl-1H-pyrido[2,1-b][1,3]benzothiazole-4-carbonyl]-L-tyrosine, RNA dependent RNA polymerase, ZINC ION | | Authors: | Tarantino, D, Mastrangelo, E, Milani, M. | | Deposit date: | 2016-03-10 | | Release date: | 2016-10-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Targeting flavivirus RNA dependent RNA polymerase through a pyridobenzothiazole inhibitor.
Antiviral Res., 134, 2016
|
|
6YP7
 
 | | PSII-LHCII C2S2 supercomplex from Pisum sativum grown in high light conditions | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Grinzato, A, Albanese, P, Zanotti, G, Pagliano, C. | | Deposit date: | 2020-04-15 | | Release date: | 2020-11-25 | | Last modified: | 2020-12-02 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | High-Light versus Low-Light: Effects on Paired Photosystem II Supercomplex Structural Rearrangement in Pea Plants.
Int J Mol Sci, 21, 2020
|
|
3MUP
 
 | | cIAP1-BIR3 domain in complex with the Smac-mimetic compound Smac037 | | Descriptor: | (3S,6S,7R,9aS)-6-{[(2S)-2-aminobutanoyl]amino}-7-(2-aminoethyl)-N-(diphenylmethyl)-5-oxooctahydro-1H-pyrrolo[1,2-a]azepine-3-carboxamide, Baculoviral IAP repeat-containing protein 2, ZINC ION | | Authors: | Cossu, F, Malvezzi, F, Canevari, G, Milani, M. | | Deposit date: | 2010-05-03 | | Release date: | 2010-11-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Recognition of Smac-mimetic compounds by the BIR domain of cIAP1
Protein Sci., 19, 2010
|
|
1KQP
 
 | | NH3-DEPENDENT NAD+ SYNTHETASE FROM BACILLUS SUBTILIS AT 1 A RESOLUTION | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, NH(3)-dependent NAD(+) synthetase, ... | | Authors: | Symersky, J, Devedjiev, Y, Moore, K, Brouillette, C, DeLucas, L. | | Deposit date: | 2002-01-07 | | Release date: | 2002-06-28 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | NH3-dependent NAD+ synthetase from Bacillus subtilis at 1 A resolution.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
6HUD
 
 | |
5O7G
 
 | |
7NMN
 
 | | Rabbit HCN4 stabilised in amphipol A8-35 | | Descriptor: | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4,Rabbit HCN4 | | Authors: | Chaves-Sanjuan, A. | | Deposit date: | 2021-02-23 | | Release date: | 2021-06-30 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Gating movements and ion permeation in HCN4 pacemaker channels.
Mol.Cell, 81, 2021
|
|
7ZH7
 
 | |
5FAE
 
 | | N184K pathological variant of gelsolin domain 2 (trigonal form) | | Descriptor: | CALCIUM ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Boni, F, Milani, M, Ricagno, S, Bolognesi, M, de Rosa, M. | | Deposit date: | 2015-12-11 | | Release date: | 2016-10-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular basis of a novel renal amyloidosis due to N184K gelsolin variant.
Sci Rep, 6, 2016
|
|
5FAF
 
 | | N184K pathological variant of gelsolin domain 2 (orthorhombic form) | | Descriptor: | ACETATE ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Boni, F, Milani, M, Ricagno, s, Bolognesi, M, de Rosa, M. | | Deposit date: | 2015-12-11 | | Release date: | 2016-10-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Molecular basis of a novel renal amyloidosis due to N184K gelsolin variant.
Sci Rep, 6, 2016
|
|
