1ZKR
 
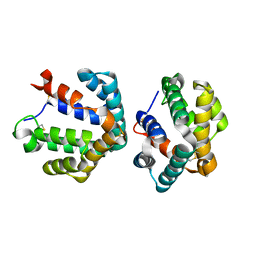 | | Crystal structure of the major cat allergen Fel d 1 (1+2) | | Descriptor: | Major allergen I polypeptide, fused chain 1, chain 2 | | Authors: | Kaiser, L, Cirkovic Velickovic, T, Adedoyin, J, Thunberg, S, Gronlund, H, Gafvelin, G, van Hage, M, Achour, A. | | Deposit date: | 2005-05-04 | | Release date: | 2006-05-16 | | Last modified: | 2017-07-26 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal structure of the major cat allergen Fel d 1 (1+2)
To be Published
|
|
1GNS
 
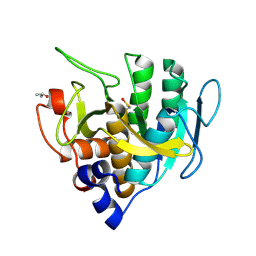 | | SUBTILISIN BPN' | | Descriptor: | ACETONE, SUBTILISIN BPN' | | Authors: | Almog, O, Gallagher, D.T, Ladner, J.E, Strausberg, S, Alexander, P. | | Deposit date: | 2001-10-06 | | Release date: | 2002-06-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of Thermostability. Analysis of Stabilizing Mutations in Subtilisin Bpn'.
J.Biol.Chem., 277, 2002
|
|
2YEN
 
 | | Solution structure of the skeletal muscle and neuronal voltage gated sodium channel antagonist mu-conotoxin CnIIIC | | Descriptor: | Mu-conotoxin CnIIIC | | Authors: | Favreau, P, Benoit, E, Hocking, H.G, Carlier, L, D'hoedt, D, Leipold, E, Markgraf, R, Schlumberger, S, Cordova, M.A, Gaertner, H, Paolini-Bertrand, M, Hartley, O, Tytgat, J, Heinemann, S.H, Bertrand, D, Boelens, R, Stocklin, R, Molgo, J. | | Deposit date: | 2011-03-28 | | Release date: | 2012-02-08 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | A Novel Mu-Conopeptide, Cniiic, Exerts Potent and Preferential Inhibition of Na(V) 1.2/1.4 Channels and Blocks Neuronal Nicotinic Acetylcholine Receptors.
Br.J.Pharmacol., 166, 2012
|
|
2XRQ
 
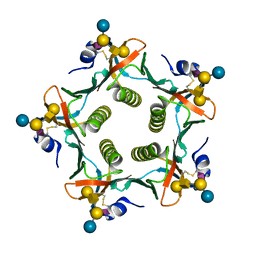 | | Crystal structures exploring the origins of the broader specificity of escherichia coli heat-labile enterotoxin compared to cholera toxin | | Descriptor: | HEAT-LABILE ENTEROTOXIN B CHAIN, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose-(1-4)-[N-acetyl-alpha-neuraminic acid-(2-3)]beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Holmner, A, Mackenzie, A, Okvist, M, Jansson, L, Lebens, M, Teneberg, S, Krengel, U. | | Deposit date: | 2010-09-19 | | Release date: | 2010-12-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures Exploring the Origins of the Broader Specificity of Escherichia Coli Heat-Labile Enterotoxin Compared to Cholera Toxin
J.Mol.Biol., 406, 2011
|
|
2XRS
 
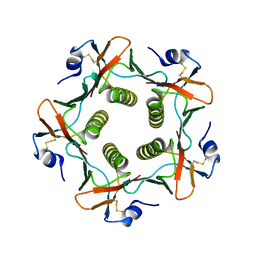 | | Crystal structures exploring the origins of the broader specificity of escherichia coli heat-labile enterotoxin compared to cholera toxin | | Descriptor: | HEAT-LABILE ENTEROTOXIN B CHAIN, beta-D-galactopyranose, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Holmner, A, Mackenzie, A, Okvist, M, Jansson, L, Lebens, M, Teneberg, S, Krengel, U. | | Deposit date: | 2010-09-20 | | Release date: | 2010-12-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal Structures Exploring the Origins of the Broader Specificity of Escherichia Coli Heat-Labile Enterotoxin Compared to Cholera Toxin
J.Mol.Biol., 406, 2011
|
|
2WV6
 
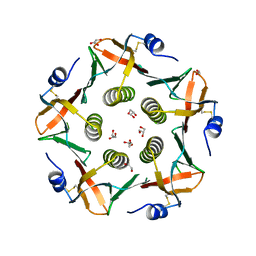 | | Crystal structure of the cholera toxin-like B-subunit from Citrobacter freundii to 1.9 angstrom | | Descriptor: | 1,2-ETHANEDIOL, CFXB, GLYCEROL | | Authors: | Jansson, L, Lebens, M, Imberty, A, Varrot, A, Teneberg, S. | | Deposit date: | 2009-10-15 | | Release date: | 2009-10-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.895 Å) | | Cite: | Carbohydrate Binding Specificities and Crystal Structure of the Cholera Toxin-Like B-Subunit from Citrobacter Freundii.
Biochimie, 92, 2010
|
|
3H79
 
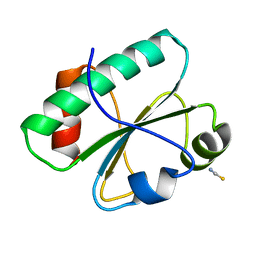 | | Crystal structure of Trypanosoma cruzi thioredoxin-like hypothetical protein Q4DV70 | | Descriptor: | THIOCYANATE ION, Thioredoxin-like protein | | Authors: | Santos, C.R, Fessel, M.R, Vieira, L.C, Krieger, M.A, Goldenberg, S, Guimaraes, B.G, Zanchin, N.I.T, Barbosa, J.A.R.G. | | Deposit date: | 2009-04-24 | | Release date: | 2009-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of Trypanosoma cruzi thioredoxin-like hypothetical protein Q4DV70
TO BE PUBLISHED
|
|
2P5L
 
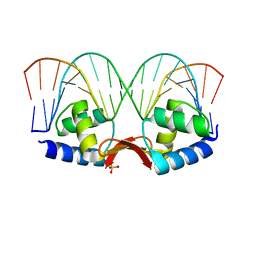 | | Crystal structure of a dimer of N-terminal domains of AhrC in complex with an 18bp DNA operator site | | Descriptor: | Arginine repressor, DNA (5'-D(*DCP*DAP*DTP*DGP*DAP*DAP*DTP*DAP*DAP*DAP*DAP*DAP*DTP*DTP*DCP*DAP*DAP*DG)-3'), DNA (5'-D(*DCP*DTP*DTP*DGP*DAP*DAP*DTP*DTP*DTP*DTP*DTP*DAP*DTP*DTP*DCP*DAP*DTP*DG)-3'), ... | | Authors: | Garnett, J.A, Marincs, F, Baumberg, S, Stockley, P.G, Phillips, S.E.V. | | Deposit date: | 2007-03-15 | | Release date: | 2008-03-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure and function of the arginine repressor-operator complex from Bacillus subtilis.
J.Mol.Biol., 379, 2008
|
|
2P5M
 
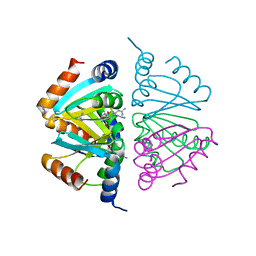 | | C-terminal domain hexamer of AhrC bound with L-arginine | | Descriptor: | ARGININE, Arginine repressor | | Authors: | Garnett, J.A, Baumberg, S, Stockley, P.G, Phillips, S.E.V. | | Deposit date: | 2007-03-15 | | Release date: | 2007-10-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of the C-terminal effector-binding domain of AhrC bound to its corepressor L-arginine.
Acta Crystallogr.,Sect.F, 63, 2007
|
|
1XBU
 
 | | Streptomyces griseus aminopeptidase complexed with p-iodo-D-phenylalanine | | Descriptor: | Aminopeptidase, CALCIUM ION, P-IODO-D-PHENYLALANINE, ... | | Authors: | Reiland, V, Gilboa, R, Spungin-Bialik, A, Schomburg, D, Shoham, Y, Blumberg, S, Shoham, G. | | Deposit date: | 2004-08-31 | | Release date: | 2005-10-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Streptomyces griseus aminopeptidase complexed with p-iodo-D-phenylalanine
To be Published
|
|
1XRY
 
 | | Crystal structure of Aeromonas proteolytica aminopeptidase in complex with bestatin | | Descriptor: | 2-(3-AMINO-2-HYDROXY-4-PHENYL-BUTYRYLAMINO)-4-METHYL-PENTANOIC ACID, Bacterial leucyl aminopeptidase, ZINC ION | | Authors: | Gilboa, R, Rondeau, J.-M, Blumberg, S, Tarnus, C, Shoham, G. | | Deposit date: | 2004-10-17 | | Release date: | 2005-09-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Interactions of Streptomyces griseus Aminopeptidase and Aeromonas proteolytica Aminopeptidase with Bestatin. Structural analysis of homologous enzymes with different binding modes.
To be Published
|
|
1TF8
 
 | | Streptomyces griseus aminopeptidase complexed with L-tryptophan | | Descriptor: | Aminopeptidase, CALCIUM ION, TRYPTOPHAN, ... | | Authors: | Reiland, V, Gilboa, R, Spungin-Bialik, A, Schomburg, D, Shoham, Y, Blumberg, S, Shoham, G. | | Deposit date: | 2004-05-27 | | Release date: | 2005-05-31 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Binding of inhibitory aromatic amino acids to Streptomyces griseus aminopeptidase.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1TKF
 
 | | Streptomyces griseus aminopeptidase complexed with D-tryptophan | | Descriptor: | Aminopeptidase, CALCIUM ION, D-TRYPTOPHAN, ... | | Authors: | Reiland, V, Gilboa, R, Spungin-Bialik, A, Schomburg, D, Shoham, Y, Blumberg, S, Shoham, G. | | Deposit date: | 2004-06-08 | | Release date: | 2005-06-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Interactions of D Amino Acids with Streptomyces griseus Aminopeptidase
To be Published
|
|
2V3L
 
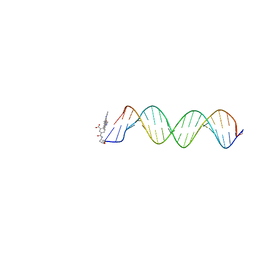 | | Orientational and dynamical heterogeneity of Rhodamine 6G terminally attached to a DNA helix | | Descriptor: | 5'-D(*CP*AP*AP*AP*GP*CP*GP*CP*CP*AP *TP*TP*CP*GP*CP*CP*AP*TP*TP*C)-3', 5'-D(*GP*AP*AP*TP*GP*GP*CP*GP*AP*AP *TP*GP*GP*CP*GP*CP*TP*TP*TP*G)-3', RHODAMINE 6G | | Authors: | Neubauer, H, Gaiko, N, Berger, S, Schaffer, J, Eggeling, C, Tuma, J, Verdier, L, Seidel, C.A.M, Griesinger, C, Volkmer, A. | | Deposit date: | 2007-06-18 | | Release date: | 2007-10-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Orientational and Dynamical Heterogeneity of Rhodamine 6G Terminally Attached to a DNA Helix Revealed by NMR and Single-Molecule Fluorescence Spectroscopy.
J.Am.Chem.Soc., 129, 2007
|
|
1TZS
 
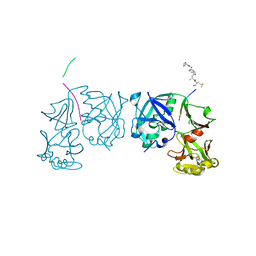 | | Crystal Structure of an activation intermediate of Cathepsin E | | Descriptor: | 23-mer peptide from PelB-IgG kappa light chain fusion protein, Cathepsin E, activation peptide from Cathepsin E | | Authors: | Ostermann, N, Gerhartz, B, Worpenberg, S, Trappe, J, Eder, J. | | Deposit date: | 2004-07-12 | | Release date: | 2005-07-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of an activation intermediate of cathepsin e
J.Mol.Biol., 342, 2004
|
|
2P5K
 
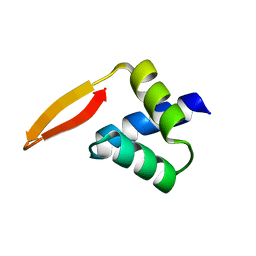 | | Crystal structure of the N-terminal domain of AhrC | | Descriptor: | Arginine repressor | | Authors: | Garnett, J.A, Baumberg, S, Stockley, P.G, Phillips, S.E.V. | | Deposit date: | 2007-03-15 | | Release date: | 2007-10-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | A high-resolution structure of the DNA-binding domain of AhrC, the arginine repressor/activator protein from Bacillus subtilis.
Acta Crystallogr.,Sect.F, 63, 2007
|
|
1EV8
 
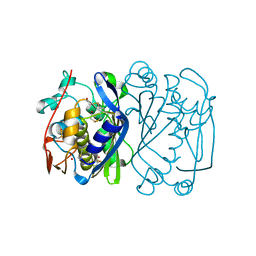 | | CRYSTAL STRUCTURE ANALYSIS OF CYS167 MUTANT OF ESCHERICHIA COLI | | Descriptor: | THYMIDYLATE SYNTHASE | | Authors: | Phan, J, Mahdavian, E, Nivens, M.C, Minor, W, Berger, S, Spencer, H.T, Dunlap, R.B, Lebioda, L. | | Deposit date: | 2000-04-19 | | Release date: | 2000-05-03 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Catalytic cysteine of thymidylate synthase is activated upon substrate binding.
Biochemistry, 39, 2000
|
|
1EVF
 
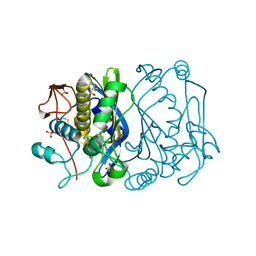 | | CRYSTAL STRUCTURE ANALYSIS OF CYS167 MUTANT OF ESCHERICHIA COLI | | Descriptor: | SULFATE ION, THYMIDYLATE SYNTHASE | | Authors: | Phan, J, Mahdavian, E, Nivens, M.C, Minor, W, Berger, S, Spencer, H.T, Dunlap, R.B, Lebioda, L. | | Deposit date: | 2000-04-19 | | Release date: | 2000-05-03 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Catalytic cysteine of thymidylate synthase is activated upon substrate binding.
Biochemistry, 39, 2000
|
|
1EV5
 
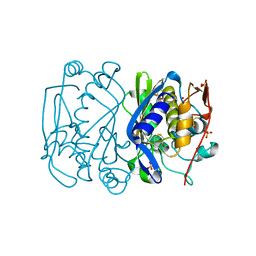 | | CRYSTAL STRUCTURE ANALYSIS OF ALA167 MUTANT OF ESCHERICHIA COLI | | Descriptor: | SULFATE ION, THYMIDYLATE SYNTHASE | | Authors: | Phan, J, Mahdavian, E, Nivens, M.C, Minor, W, Berger, S, Spencer, H.T, Dunlap, R.B, Lebioda, L. | | Deposit date: | 2000-04-19 | | Release date: | 2000-05-03 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Catalytic cysteine of thymidylate synthase is activated upon substrate binding.
Biochemistry, 39, 2000
|
|
1EVG
 
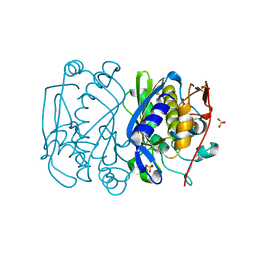 | | CRYSTAL STRUCTURE ANALYSIS OF CYS167 MUTANT OF ESCHERICHIA COLI WITH UNMODIFIED CATALYTIC CYSTEINE | | Descriptor: | SULFATE ION, THYMIDYLATE SYNTHASE | | Authors: | Phan, J, Mahdavian, E, Nivens, M.C, Minor, W, Berger, S, Spencer, H.T, Dunlap, R.B, Lebioda, L. | | Deposit date: | 2000-04-19 | | Release date: | 2000-05-03 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Catalytic cysteine of thymidylate synthase is activated upon substrate binding.
Biochemistry, 39, 2000
|
|
4AML
 
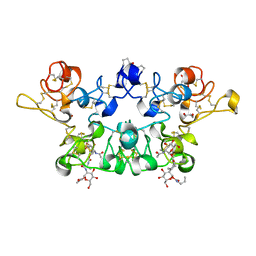 | | CRYSTAL STRUCTURE OF WHEAT GERM AGGLUTININ ISOLECTIN 1 IN COMPLEX WITH GLYCOSYLURETHAN | | Descriptor: | 2-acetamido-2-deoxy-1-O-(propylcarbamoyl)-alpha-D-glucopyranose, AGGLUTININ ISOLECTIN 1, GLYCEROL | | Authors: | Schwefel, D, Maierhofer, C, Beck, J.G, Seeberger, S, Diederichs, K, Moeller, H.M, Welte, W, Wittmann, V. | | Deposit date: | 2012-03-12 | | Release date: | 2012-04-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Basis of Multivalent Binding to Wheat Germ Agglutinin.
J.Am.Chem.Soc., 132, 2010
|
|
6YT3
 
 | | Structure of the MoStoNano fusion protein | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Molybdenum storage protein subunit alpha, ... | | Authors: | Benoit, R.M, Bierig, T, Collu, C, Engilberge, S, Olieric, V. | | Deposit date: | 2020-04-23 | | Release date: | 2020-12-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Chimeric single alpha-helical domains as rigid fusion protein connections for protein nanotechnology and structural biology.
Structure, 30, 2022
|
|
6YT1
 
 | | Mtb TMK crystal structure in complex with compound 26 | | Descriptor: | 2-ethyl-~{N}-[[4-[4-[5-methyl-2,4-bis(oxidanylidene)pyrimidin-1-yl]piperidin-1-yl]phenyl]methyl]-1,2,3,5,6,7,8,8~{a}-octahydroimidazo[1,2-a]pyridine-3-carboxamide, CITRIC ACID, Thymidylate kinase | | Authors: | Merceron, R, De Munck, S, Jian, Y, Munier-Lehmann, H, Van Calenbergh, S, Savvides, S.N. | | Deposit date: | 2020-04-23 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Endeavors towards transformation of M. tuberculosis thymidylate kinase (MtbTMPK) inhibitors into potential antimycobacterial agents.
Eur.J.Med.Chem., 206, 2020
|
|
6Z5Q
 
 | | The RSL - sulfonato-calix[8]arene complex, P3 form, acetate pH 4.0 | | Descriptor: | Fucose-binding lectin protein, GLYCEROL, beta-D-fructopyranose, ... | | Authors: | Ramberg, K, Engilberge, S, Crowley, P.B. | | Deposit date: | 2020-05-27 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Facile Fabrication of Protein-Macrocycle Frameworks.
J.Am.Chem.Soc., 143, 2021
|
|
6Z5P
 
 | | The RSL-R8 - sulfonato-calix[8]arene complex, P3 form, TRIS-HCl pH 8.5 | | Descriptor: | Fucose-binding lectin protein, GLYCEROL, SULFATE ION, ... | | Authors: | Ramberg, K, Skorek, T, Engilberge, S, Crowley, P.B. | | Deposit date: | 2020-05-27 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Facile Fabrication of Protein-Macrocycle Frameworks.
J.Am.Chem.Soc., 143, 2021
|
|
