4QQG
 
 | | Crystal structure of an N-terminal HTATIP fragment | | Descriptor: | Histone acetyltransferase KAT5, UNKNOWN ATOM OR ION | | Authors: | Liu, Y, Tempel, W, Wernimont, A.K, Dobrovetsky, E, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-06-27 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and histone binding studies of the chromo barrel domain of TIP60.
FEBS Lett., 592, 2018
|
|
4QSS
 
 | | Structure of the bromodomain of human ATPase family AAA domain-containing protein 2 (ATAD2) in complex with N-Methyl-2-pyrrolidone (NMP) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1-methylpyrrolidin-2-one, ATPase family AAA domain-containing protein 2, ... | | Authors: | Chaikuad, A, Felletar, I, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-07-06 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based approaches towards identification of fragments for the low-druggability ATAD2 bromodomain
MedChemComm, 5, 2014
|
|
6RU8
 
 | | Crystal structure of Casein Kinase I delta (CK1d) in complex with triple phosphorylated p63 PAD3P peptide | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, Casein kinase I isoform delta, ... | | Authors: | Chaikuad, A, Tuppi, M, Gebel, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Dotsch, V, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-05-27 | | Release date: | 2020-05-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | p63 uses a switch-like mechanism to set the threshold for induction of apoptosis.
Nat.Chem.Biol., 16, 2020
|
|
6RV3
 
 | | Crystal structure of the human two pore domain potassium ion channel TASK-1 (K2P3.1) in a closed conformation with a bound inhibitor BAY 1000493 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CHOLESTEROL HEMISUCCINATE, DECYL-BETA-D-MALTOPYRANOSIDE, ... | | Authors: | Rodstrom, K.E.J, Pike, A.C.W, Zhang, W, Quigley, A, Speedman, D, Mukhopadhyay, S.M.M, Shrestha, L, Chalk, R, Venkaya, S, Bushell, S.R, Tessitore, A, Burgess-Brown, N, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-05-30 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A lower X-gate in TASK channels traps inhibitors within the vestibule.
Nature, 582, 2020
|
|
3FM8
 
 | | Crystal structure of full length centaurin alpha-1 bound with the FHA domain of KIF13B (CAPRI target) | | Descriptor: | Centaurin-alpha-1, Kinesin-like protein KIF13B, SULFATE ION, ... | | Authors: | Shen, L, Tong, Y, Tempel, W, MacKenzie, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-12-19 | | Release date: | 2009-08-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Phosphorylation-independent dual-site binding of the FHA domain of KIF13 mediates phosphoinositide transport via centaurin alpha1.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4RXJ
 
 | | crystal structure of WHSC1L1-PWWP2 | | Descriptor: | Histone-lysine N-methyltransferase NSD3, UNKNOWN ATOM OR ION | | Authors: | Qin, S, Tempel, W, Dong, A, Li, Y, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-12-11 | | Release date: | 2015-01-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Histone and DNA binding ability studies of the NSD subfamily of PWWP domains.
Biochem.Biophys.Res.Commun., 569, 2021
|
|
3F2O
 
 | | Crystal Structure of human splA/ryanodine receptor domain and SOCS box containing 1 (SPSB1) in complex with a 20-residue VASA peptide | | Descriptor: | 20-mer peptide from ATP-dependent RNA helicase vasa, SPRY domain-containing SOCS box protein 1 | | Authors: | Filippakopoulos, P, Sharpe, T, Keates, T, Murray, J.W, Savitsky, P, Roos, A, Pike, A.C.W, Von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Bullock, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-10-30 | | Release date: | 2008-12-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for Par-4 recognition by the SPRY domain- and SOCS box-containing proteins SPSB1, SPSB2, and SPSB4.
J.Mol.Biol., 401, 2010
|
|
3FDT
 
 | | Crystal structure of the complex of human chromobox homolog 5 (CBX5) with H3K9(me)3 peptide | | Descriptor: | Chromobox protein homolog 5, H3K9(me)3 peptide | | Authors: | Amaya, M.F, Ravichandran, M, Loppnau, P, Kozieradzki, I, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Bountra, C, Bochkarev, A, Min, J, Ouyang, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-11-26 | | Release date: | 2009-01-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Recognition and specificity determinants of the human cbx chromodomains.
J.Biol.Chem., 286, 2011
|
|
6T1J
 
 | | Crystal structure of MLLT1 (ENL) YEATS domain in complexed with piperazine-urea derivative 2 | | Descriptor: | 1,2-ETHANEDIOL, Protein ENL, ~{N}-[[4-(pyrrolidin-1-ylmethyl)phenyl]methyl]-4-thiophen-2-ylcarbonyl-piperazine-1-carboxamide | | Authors: | Chaikuad, A, Heidenreich, D, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Fedorov, O, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-10-04 | | Release date: | 2019-11-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural Insights into Interaction Mechanisms of Alternative Piperazine-urea YEATS Domain Binders in MLLT1.
Acs Med.Chem.Lett., 10, 2019
|
|
6QU1
 
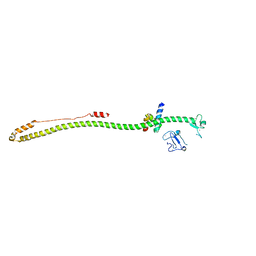 | | Crystal structure of the KAP1 RBCC domain in complex with the SMARCAD1 CUE1 domain at 3.7 angstrom resolution. | | Descriptor: | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A containing DEAD/H box 1, Transcription intermediary factor 1-beta,Transcription intermediary factor 1-beta, ZINC ION | | Authors: | Newman, J.A, Aitkenhead, H, Gavard, A, Lim, M, Williams, H.L, Svejstrup, J.Q, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2019-02-26 | | Release date: | 2019-07-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | A Ubiquitin-Binding Domain that Binds a Structural Fold Distinct from that of Ubiquitin.
Structure, 2019
|
|
6SZM
 
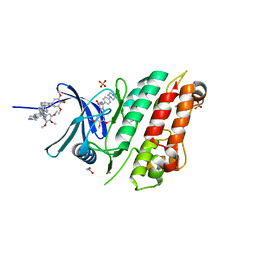 | | Crystal structure of the ACVR1 (ALK2) kinase in complex with the compound M4K2009 | | Descriptor: | 1,2-ETHANEDIOL, 1-[4-[4-methyl-5-(3,4,5-trimethoxyphenyl)pyridin-3-yl]phenyl]piperazine, AMMONIUM ION, ... | | Authors: | Adamson, R.J, Williams, E.P, Smil, D, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2019-10-02 | | Release date: | 2019-10-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Leveraging an Open Science Drug Discovery Model to Develop CNS-Penetrant ALK2 Inhibitors for the Treatment of Diffuse Intrinsic Pontine Glioma.
J.Med.Chem., 63, 2020
|
|
3FEG
 
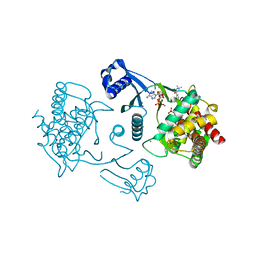 | | Crystal structure of human choline kinase beta in complex with phosphorylated hemicholinium-3 and adenosine nucleotide | | Descriptor: | (2S)-2-[4'-({dimethyl[2-(phosphonooxy)ethyl]ammonio}acetyl)biphenyl-4-yl]-2-hydroxy-4,4-dimethylmorpholin-4-ium, ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Hong, B.S, Tempel, W, Rabeh, W.M, MacKenzie, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H.W, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-11-29 | | Release date: | 2008-12-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.302 Å) | | Cite: | Crystal structures of human choline kinase isoforms in complex with hemicholinium-3: single amino acid near the active site influences inhibitor sensitivity.
J.Biol.Chem., 285, 2010
|
|
6T1L
 
 | | Crystal structure of MLLT1 (ENL) YEATS domain in complexed with piperazine-urea derivative 3 | | Descriptor: | 1,2-ETHANEDIOL, Protein ENL, ~{N}-[[4-(diethylaminomethyl)phenyl]methyl]-4-pyrimidin-2-yl-piperazine-1-carboxamide | | Authors: | Chaikuad, A, Heidenreich, D, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Fedorov, O, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-10-04 | | Release date: | 2019-11-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insights into Interaction Mechanisms of Alternative Piperazine-urea YEATS Domain Binders in MLLT1.
Acs Med.Chem.Lett., 10, 2019
|
|
3FVY
 
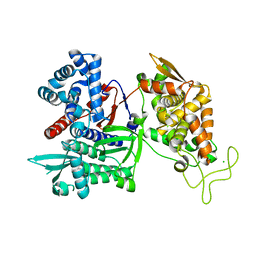 | | Crystal structure of human Dipeptidyl Peptidase III | | Descriptor: | CHLORIDE ION, Dipeptidyl-peptidase 3, MAGNESIUM ION, ... | | Authors: | Dong, A, Dobrovetsky, E, Seitova, A, Duncan, B, Crombet, L, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bochkarev, A, Cossar, D, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-01-16 | | Release date: | 2009-02-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Entropy-driven binding of opioid peptides induces a large domain motion in human dipeptidyl peptidase III.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
6RRK
 
 | | Crystal structure of the central region of human cohesin subunit STAG1 in complex with RAD21 peptide | | Descriptor: | Cohesin subunit SA-1, Double-strand-break repair protein rad21 homolog | | Authors: | Newman, J.A, katis, V.L, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2019-05-20 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | STAG1 vulnerabilities for exploiting cohesin synthetic lethality in STAG2-deficient cancers.
Life Sci Alliance, 3, 2020
|
|
6T1N
 
 | | Crystal structure of MLLT1 (ENL) YEATS domain in complexed with benzimidazole-amide derivative 5 | | Descriptor: | 1,2-ETHANEDIOL, 4-chloranyl-~{N}-[2-(piperidin-1-ylmethyl)-3~{H}-benzimidazol-5-yl]benzamide, Protein ENL | | Authors: | Chaikuad, A, Heidenreich, D, Moustakim, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Fedorov, O, Brennan, P.E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-10-04 | | Release date: | 2019-11-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Insights into Interaction Mechanisms of Alternative Piperazine-urea YEATS Domain Binders in MLLT1.
Acs Med.Chem.Lett., 10, 2019
|
|
3FY2
 
 | | Human EphA3 Kinase and Juxtamembrane Region Bound to Substrate KQWDNYEFIW | | Descriptor: | Ephrin type-A receptor 3, peptide substrate | | Authors: | Davis, T, Walker, J.R, Mackenzie, F, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-01-21 | | Release date: | 2009-02-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural recognition of an optimized substrate for the ephrin family of receptor tyrosine kinases.
Febs J., 276, 2009
|
|
6T6B
 
 | | Crystal structure of PPARgamma in complex with compound 16 (MF27) | | Descriptor: | (2~{R})-2-[[6-[(2,4-dichlorophenyl)sulfonylamino]-1,3-benzothiazol-2-yl]sulfanyl]octanoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Chaikuad, A, Ni, X, Hanke, T, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Merk, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-10-18 | | Release date: | 2019-12-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A Selective Modulator of Peroxisome Proliferator-Activated Receptor gamma with an Unprecedented Binding Mode.
J.Med.Chem., 63, 2020
|
|
4RYL
 
 | | Human Protein Arginine Methyltransferase 3 in complex with 1-isoquinolin-6-yl-3-[2-oxo-2-(pyrrolidin-1-yl)ethyl]urea | | Descriptor: | 1-isoquinolin-6-yl-3-[2-oxo-2-(pyrrolidin-1-yl)ethyl]urea, PRMT3 protein, UNKNOWN ATOM OR ION | | Authors: | Dong, A, Dobrovetsky, E, Kaniskan, H.U, Szewczyk, M, Yu, Z, Eram, M.S, Yang, X, Schmidt, K, Luo, X, Dai, M, He, F, Zang, I, Lin, Y, Kennedy, S, Li, F, Tempel, W, Smil, D, Min, S.J, Landon, M, Lin-Jones, J, Huang, X.P, Roth, B.L, Schapira, M, Atadja, P, Barsyte-Lovejoy, D, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Brown, P.J, Zhao, K, Jin, J, Vedadi, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-12-15 | | Release date: | 2015-02-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Potent, Selective and Cell-Active Allosteric Inhibitor of Protein Arginine Methyltransferase 3 (PRMT3).
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4RG2
 
 | | Tudor Domain of Tumor suppressor p53BP1 with small molecule ligand | | Descriptor: | 1,2-ETHANEDIOL, 3-bromo-N-[3-(tert-butylamino)propyl]benzamide, Tumor suppressor p53-binding protein 1, ... | | Authors: | Dong, A, Mader, P, James, L, Perfetti, M, Tempel, W, Frye, S, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-09-29 | | Release date: | 2014-10-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Identification of a fragment-like small molecule ligand for the methyl-lysine binding protein, 53BP1.
ACS Chem. Biol., 10, 2015
|
|
4QST
 
 | | Structure of the bromodomain of human ATPase family AAA domain-containing protein 2 (ATAD2) in complex with 1-methylquinolin 2-one | | Descriptor: | 1,2-ETHANEDIOL, 1-METHYLQUINOLIN-2(1H)-ONE, ATPase family AAA domain-containing protein 2, ... | | Authors: | Chaikuad, A, Felletar, I, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-07-06 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure-based approaches towards identification of fragments for the low-druggability ATAD2 bromodomain
MedChemComm, 5, 2014
|
|
4QSP
 
 | | Structure of the bromodomain of human ATPase family AAA domain-containing protein 2 (ATAD2) in complex with acetyl-lysine | | Descriptor: | 1,2-ETHANEDIOL, ATPase family AAA domain-containing protein 2, N(6)-ACETYLLYSINE, ... | | Authors: | Chaikuad, A, Felletar, I, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-07-06 | | Release date: | 2014-07-23 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-based approaches towards identification of fragments for the low-druggability ATAD2 bromodomain
MedChemComm, 5, 2014
|
|
4QSV
 
 | | Structure of the bromodomain of human ATPase family AAA domain-containing protein 2 (ATAD2) in complex with thymidine | | Descriptor: | 1,2-ETHANEDIOL, ATPase family AAA domain-containing protein 2, SULFATE ION, ... | | Authors: | Chaikuad, A, Felletar, I, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-07-06 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-based approaches towards identification of fragments for the low-druggability ATAD2 bromodomain
MedChemComm, 5, 2014
|
|
4TXC
 
 | | Crystal Structure of DAPK1 kinase domain in complex with a small molecule inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 4-(3-{3-[(R)-{[2-(dimethylamino)ethyl]amino}(hydroxy)methyl]phenyl}imidazo[1,2-b]pyridazin-6-yl)-2-methoxyphenol, Death-associated protein kinase 1 | | Authors: | Sorrell, F.J, Krojer, T, Wilbek, T.S, Skovgaard, T, Berthelsen, J, Dixon-Clarke, S, Chalk, R, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Stromgaard, K, Knapp, S. | | Deposit date: | 2014-07-03 | | Release date: | 2014-07-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | Identification and characterization of a small-molecule inhibitor of death-associated protein kinase 1.
Chembiochem, 16, 2015
|
|
4QQ6
 
 | | Crystal Structure of tudor domain of SMN1 in complex with a small organic molecule | | Descriptor: | 4-methyl-2,3,4,5,6,7-hexahydrodicyclopenta[b,e]pyridin-8(1H)-imine, Survival motor neuron protein, UNKNOWN ATOM OR ION | | Authors: | Liu, Y, Tempel, W, Iqbal, A, Walker, J.R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-06-26 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A small molecule antagonist of SMN disrupts the interaction between SMN and RNAP II.
Nat Commun, 13, 2022
|
|
