3GPV
 
 | | Crystal structure of a transcriptional regulator, MerR family from Bacillus thuringiensis | | Descriptor: | Transcriptional regulator, MerR family | | Authors: | Palani, K, Kumaran, D, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-03-23 | | Release date: | 2009-04-14 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a transcriptional regulator, MerR family from Bacillus thuringiensis
To be Published
|
|
2GU1
 
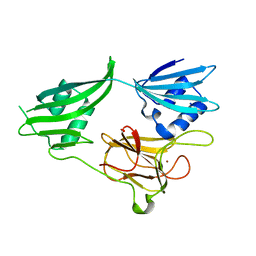 | | Crystal structure of a zinc containing peptidase from vibrio cholerae | | Descriptor: | SODIUM ION, ZINC ION, Zinc peptidase | | Authors: | Sugadev, R, Kumaran, D, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-04-28 | | Release date: | 2006-07-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a putative lysostaphin peptidase from Vibrio cholerae.
Proteins, 72, 2008
|
|
2SPC
 
 | | CRYSTAL STRUCTURE OF THE REPETITIVE SEGMENTS OF SPECTRIN | | Descriptor: | SPECTRIN | | Authors: | Yan, Y, Winograd, E, Viel, A, Cronin, T, Harrison, S.C, Branton, D. | | Deposit date: | 1994-03-01 | | Release date: | 1994-05-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the repetitive segments of spectrin.
Science, 262, 1993
|
|
2GOK
 
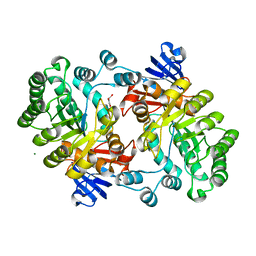 | | Crystal structure of the imidazolonepropionase from Agrobacterium tumefaciens at 1.87 A resolution | | Descriptor: | CHLORIDE ION, FE (III) ION, GLYCEROL, ... | | Authors: | Tyagi, R, Kumaran, D, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-04-13 | | Release date: | 2006-04-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | X-ray structure of imidazolonepropionase from Agrobacterium tumefaciens at 1.87 A resolution.
Proteins, 69, 2007
|
|
3GRA
 
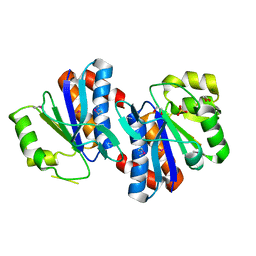 | | Crystal structure of AraC family transcriptional regulator from Pseudomonas putida | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Bagaria, A, Kumaran, D, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-03-25 | | Release date: | 2009-04-14 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of AraC family transcriptional regulator from Pseudomonas putida
To be Published
|
|
3HYO
 
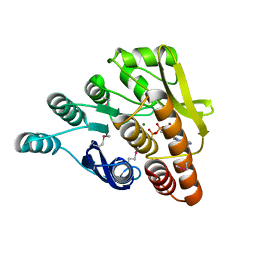 | | Crystal structure of pyridoxal kinase from Lactobacillus plantarum in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Pyridoxal kinase | | Authors: | Bagaria, A, Kumaran, D, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-06-22 | | Release date: | 2009-06-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of pyridoxal kinase from Lactobacillus plantarum in complex with ADP
To be Published
|
|
4KL1
 
 | | HCN4 CNBD in complex with cGMP | | Descriptor: | ACETATE ION, CYCLIC GUANOSINE MONOPHOSPHATE, GLYCEROL, ... | | Authors: | Lolicato, M, Arrigoni, C, Zucca, S, Nardini, M, Bucchi, A, Schroeder, I, Simmons, K, Bolognesi, M, DiFrancesco, D, Schwede, F, Fishwick, C.W.G, Johnson, A.P.K, Thiel, G, Moroni, A. | | Deposit date: | 2013-05-07 | | Release date: | 2014-04-30 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Cyclic dinucleotides bind the C-linker of HCN4 to control channel cAMP responsiveness.
Nat.Chem.Biol., 10, 2014
|
|
3GRC
 
 | |
2HAF
 
 | | Crystal structure of a putative translation repressor from Vibrio cholerae | | Descriptor: | Putative translation repressor | | Authors: | Sugadev, R, Seetharaman, J, Kumaran, D, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-06-12 | | Release date: | 2006-07-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Crystal structure of a putative translation repressor from Vibrio cholerae
To be Published
|
|
6C2W
 
 | | Crystal structure of human prothrombin mutant S101C/A470C | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, Prothrombin, ... | | Authors: | Chinnaraj, M, Chen, Z, Pelc, L, Grese, Z, Bystranowska, D, Di Cera, E, Pozzi, N. | | Deposit date: | 2018-01-09 | | Release date: | 2018-02-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (4.12 Å) | | Cite: | Structure of prothrombin in the closed form reveals new details on the mechanism of activation.
Sci Rep, 8, 2018
|
|
3R62
 
 | | Structure of complement regulator Factor H mutant, T1184R. | | Descriptor: | Complement factor H, GLYCEROL | | Authors: | Morgan, H.P, Jiang, J, Herbert, A.P, Kavanagh, D, Uhran, D, Barlow, P.N, Hannan, J.P. | | Deposit date: | 2011-03-21 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Crystallographic determination of the disease-associated T1184R variant of the complement Factor H.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3ILD
 
 | | Structure of ORF157-K57A from Acidianus filamentous virus 1 | | Descriptor: | MAGNESIUM ION, Putative uncharacterized protein | | Authors: | Goulet, A, Lichiere, J, Prangishvili, D, van Tilbeurgh, H, Cambillau, C, Campanacci, V. | | Deposit date: | 2009-08-07 | | Release date: | 2010-03-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | ORF157 from the archaeal virus Acidianus filamentous virus 1 defines a new class of nuclease
J.Virol., 84, 2010
|
|
2QXY
 
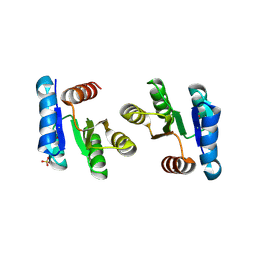 | |
2VDU
 
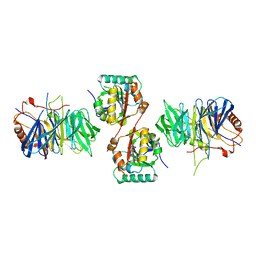 | | Structure of trm8-trm82, THE YEAST TRNA m7G methylation complex | | Descriptor: | PHOSPHATE ION, TRNA (GUANINE-N(7)-)-METHYLTRANSFERASE, TRNA (GUANINE-N(7)-)-METHYLTRANSFERASE-ASSOCIATED WD REPEAT PROTEIN TRM82 | | Authors: | Leulliot, N, Chaillet, M, Durand, D, Ulryck, N, Blondeau, K, Van Tilbeurgh, H. | | Deposit date: | 2007-10-11 | | Release date: | 2007-12-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the Yeast tRNA M7G Methylation Complex.
Structure, 16, 2008
|
|
3ILE
 
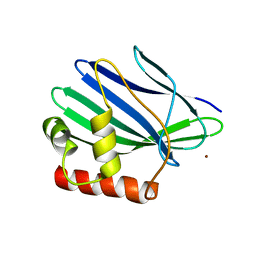 | | Crystal structure of ORF157-E86A of Acidianus filamentous virus 1 | | Descriptor: | NICKEL (II) ION, Putative uncharacterized protein | | Authors: | Goulet, A, Lichiere, J, Prangishvili, D, van Tilbeurgh, H, Cambillau, C, Campanacci, V. | | Deposit date: | 2009-08-07 | | Release date: | 2010-03-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | ORF157 from the archaeal virus Acidianus filamentous virus 1 defines a new class of nuclease
J.Virol., 84, 2010
|
|
2HFV
 
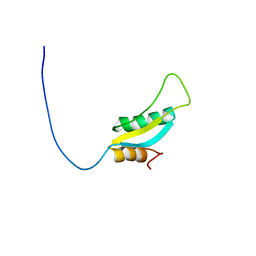 | | Solution NMR Structure of Protein RPA1041 from Pseudomonas aeruginosa. Northeast Structural Genomics Consortium Target PaT90. | | Descriptor: | Hypothetical Protein RPA1041 | | Authors: | Eletsky, A, Atreya, H.S, Liu, G, Sukumaran, D, Garcia, M, Yee, A, Arrowsmith, C, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-06-26 | | Release date: | 2006-07-25 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of Pseudomonas aeruginosa Hypothetical Protein RPA1041
TO BE PUBLISHED
|
|
2UW1
 
 | | Ivy Desaturase Structure | | Descriptor: | (3R)-3-HYDROXY-5,5-DIMETHYLHEXANOIC ACID, FE (III) ION, PLASTID DELTA4 MULTIFUNCTIONAL ACYL-ACYL CARRIER PROTEIN DESATURASE, ... | | Authors: | Guy, J.E, Whittle, E, Kumaran, D, Lindqvist, Y, Shanklin, J. | | Deposit date: | 2007-03-15 | | Release date: | 2007-05-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Crystal Structure of the Ivy {Delta}4-16:0-Acp Desaturase Reveals Structural Details of the Oxidized Active Site and Potential Determinants of Regioselectivity.
J.Biol.Chem., 282, 2007
|
|
3OVG
 
 | | The crystal structure of an amidohydrolase from Mycoplasma synoviae with Zn ion bound | | Descriptor: | PHOSPHATE ION, ZINC ION, amidohydrolase | | Authors: | Zhang, Z, Kumaran, D, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-09-16 | | Release date: | 2010-10-13 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.059 Å) | | Cite: | The crystal structure of an amidohydrolase from Mycoplasma synoviae with Zn ion bound
To be Published
|
|
3P26
 
 | | Crystal structure of S. cerevisiae Hbs1 protein (apo-form), a translational GTPase involved in RNA quality control pathways and interacting with Dom34/Pelota | | Descriptor: | Elongation factor 1 alpha-like protein | | Authors: | van den Elzen, A, Henri, J, Lazar, N, Gas, M.E, Durand, D, Lacroute, F, Nicaise, M, van Tilbeurgh, H, Sraphin, B, Graille, M, Paris-Sud Yeast Structural Genomics (YSG) | | Deposit date: | 2010-10-01 | | Release date: | 2010-11-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Dissection of Dom34-Hbs1 reveals independent functions in two RNA quality control pathways.
Nat.Struct.Mol.Biol., 17, 2010
|
|
6UAN
 
 | | B-Raf:14-3-3 complex | | Descriptor: | 14-3-3 zeta, Serine/threonine-protein kinase B-raf | | Authors: | Kondo, Y, Ognjenovic, J, Banerjee, S, Karandur, D, Merk, A, Kulhanek, K, Wong, K, Roose, J.P, Subramaniam, S, Kuriyan, J. | | Deposit date: | 2019-09-11 | | Release date: | 2019-09-25 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structure of a dimeric B-Raf:14-3-3 complex reveals asymmetry in the active sites of B-Raf kinases.
Science, 366, 2019
|
|
3P27
 
 | | Crystal structure of S. cerevisiae Hbs1 protein (GDP-bound form), a translational GTPase involved in RNA quality control pathways and interacting with Dom34/Pelota | | Descriptor: | Elongation factor 1 alpha-like protein, GUANOSINE-5'-DIPHOSPHATE | | Authors: | van den Elzen, A, Henri, J, Lazar, N, Gas, M.E, Durand, D, Lacroute, F, Nicaise, M, van Tilbeurgh, H, Sraphin, B, Graille, M, Paris-Sud Yeast Structural Genomics (YSG) | | Deposit date: | 2010-10-01 | | Release date: | 2010-11-17 | | Last modified: | 2012-03-14 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Dissection of Dom34-Hbs1 reveals independent functions in two RNA quality control pathways.
Nat.Struct.Mol.Biol., 17, 2010
|
|
5AFH
 
 | | alpha7-AChBP in complex with lobeline | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINE-BINDING PROTEIN, NEURONAL ACETYLCHOLINE RECEPTOR SUBUNIT ALPHA-7, ... | | Authors: | Spurny, R, Debaveye, S, Farinha, A, Veys, K, Gossas, T, Atack, J, Bertrand, D, Kemp, J, Vos, A, Danielson, U.H, Tresadern, G, Ulens, C. | | Deposit date: | 2015-01-22 | | Release date: | 2015-05-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular Blueprint of Allosteric Binding Sites in a Homologue of the Agonist-Binding Domain of the Alpha7 Nicotinic Acetylcholine Receptor.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5AFL
 
 | | alpha7-AChBP in complex with lobeline and fragment 3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINE-BINDING PROTEIN, NEURONAL ACETYLCHOLINE RECEPTOR SUBUNIT ALPHA-7, ... | | Authors: | Spurny, R, Debaveye, S, Farinha, A, Veys, K, Gossas, T, Atack, J, Bertrand, D, Kemp, J, Vos, A, Danielson, U.H, Tresadern, G, Ulens, C. | | Deposit date: | 2015-01-22 | | Release date: | 2015-05-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.385 Å) | | Cite: | Molecular Blueprint of Allosteric Binding Sites in a Homologue of the Agonist-Binding Domain of the Alpha7 Nicotinic Acetylcholine Receptor.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5AFN
 
 | | alpha7-AChBP in complex with lobeline and fragment 5 | | Descriptor: | (4R)-4-(2-phenylethyl)pyrrolidin-2-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINE-BINDING PROTEIN, ... | | Authors: | Spurny, R, Debaveye, S, Farinha, A, Veys, K, Gossas, T, Atack, J, Bertrand, D, Kemp, J, Vos, A, Danielson, U.H, Tresadern, G, Ulens, C. | | Deposit date: | 2015-01-22 | | Release date: | 2015-05-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.149 Å) | | Cite: | Molecular Blueprint of Allosteric Binding Sites in a Homologue of the Agonist-Binding Domain of the Alpha7 Nicotinic Acetylcholine Receptor.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5AFJ
 
 | | alpha7-AChBP in complex with lobeline and fragment 1 | | Descriptor: | (3S)-6-(4-bromophenyl)-3-hydroxy-1,3-dimethyl-2,3-dihydropyridin-4(1H)-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINE-BINDING PROTEIN, ... | | Authors: | Spurny, R, Debaveye, S, Farinha, A, Veys, K, Gossas, T, Atack, J, Bertrand, D, Kemp, J, Vos, A, Danielson, U.H, Tresadern, G, Ulens, C. | | Deposit date: | 2015-01-22 | | Release date: | 2015-05-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular Blueprint of Allosteric Binding Sites in a Homologue of the Agonist-Binding Domain of the Alpha7 Nicotinic Acetylcholine Receptor.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
