3OQO
 
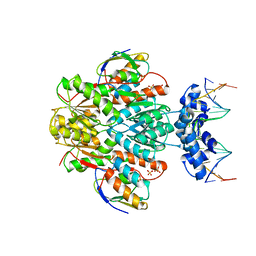 | | Ccpa-hpr-ser46p-syn cre | | Descriptor: | 5'-D(*CP*TP*GP*AP*AP*AP*GP*CP*GP*CP*TP*AP*AP*CP*AP*G)-3', 5'-D(*CP*TP*GP*TP*TP*AP*GP*CP*GP*CP*TP*TP*TP*CP*AP*G)-3', Catabolite control protein A, ... | | Authors: | schumacher, M.A, Sprehe, M, Bartholomae, M, Hillen, W, Brennan, R.G. | | Deposit date: | 2010-09-03 | | Release date: | 2011-10-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Structures of carbon catabolite protein A-(HPr-Ser46-P) bound to diverse catabolite response element sites reveal the basis for high-affinity binding to degenerate DNA operators.
Nucleic Acids Res., 39, 2011
|
|
3RNJ
 
 | | Crystal structure of the SH3 domain from IRSp53 (BAIAP2) | | Descriptor: | 1,2-ETHANEDIOL, Brain-specific angiogenesis inhibitor 1-associated protein 2, ISOPROPYL ALCOHOL, ... | | Authors: | Simister, P.C, Barilari, M, Muniz, J.R.C, Dente, L, Knapp, S, von Delft, F, Filippakopoulos, P, Vollmar, M, Chaikuad, A, Raynor, J, Tregubova, A, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Feller, S.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-04-22 | | Release date: | 2011-05-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the SH3 domain from IRSp53 (BAIAP2)
To be Published
|
|
2Q6F
 
 | | Crystal structure of infectious bronchitis virus (IBV) main protease in complex with a Michael acceptor inhibitor N3 | | Descriptor: | Infectious bronchitis virus (IBV) main protease, N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]ALANYL-L-VALYL-N~1~-((1R,2Z)-4-(BENZYLOXY)-4-OXO-1-{[(3R)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE | | Authors: | Xue, X.Y, Yang, H.T, Xue, F, Bartlam, M, Rao, Z.H. | | Deposit date: | 2007-06-05 | | Release date: | 2008-02-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of two coronavirus main proteases: implications for substrate binding and antiviral drug design.
J.Virol., 82, 2008
|
|
2QC3
 
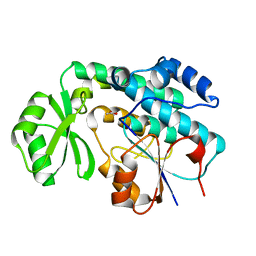 | | Crystal structure of MCAT from Mycobacterium tuberculosis | | Descriptor: | ACETIC ACID, Malonyl CoA-acyl carrier protein transacylase | | Authors: | Li, Z, Huang, Y, Ge, J, Bartlam, M, Wang, H, Rao, Z. | | Deposit date: | 2007-06-19 | | Release date: | 2007-08-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Crystal Structure of MCAT from Mycobacterium tuberculosis Reveals Three New Catalytic Models.
J.Mol.Biol., 371, 2007
|
|
2R2Z
 
 | | The crystal structure of a hemolysin domain from Enterococcus faecalis V583 | | Descriptor: | Hemolysin, ZINC ION | | Authors: | Zhang, R, Tan, K, Zhou, M, Bargassa, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-08-28 | | Release date: | 2007-09-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The crystal structure of a hemolysin domain from Enterococcus faecalis V583.
To be Published
|
|
3CHG
 
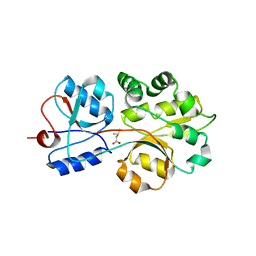 | | The compatible solute-binding protein OpuAC from Bacillus subtilis in complex with DMSA | | Descriptor: | (dimethyl-lambda~4~-sulfanyl)acetic acid, Glycine betaine-binding protein | | Authors: | Smits, S.H.J, Hoing, M, Lecher, J, Jebbar, M, Schmitt, L, Bremer, E. | | Deposit date: | 2008-03-09 | | Release date: | 2008-08-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Compatible-Solute-Binding Protein OpuAC from Bacillus subtilis: Ligand Binding, Site-Directed Mutagenesis, and Crystallographic Studies
J.Bacteriol., 190, 2008
|
|
3DJU
 
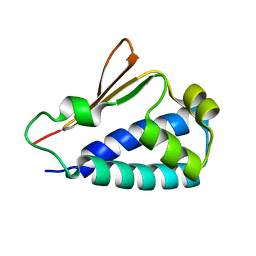 | | Crystal structure of human BTG2 | | Descriptor: | Protein BTG2 | | Authors: | Yang, X, Morita, M, Wang, H, Suzuki, T, Bartlam, M, Yamamoto, T. | | Deposit date: | 2008-06-24 | | Release date: | 2008-11-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal structures of human BTG2 and mouse TIS21 involved in suppression of CAF1 deadenylase activity
Nucleic Acids Res., 36, 2008
|
|
3IWF
 
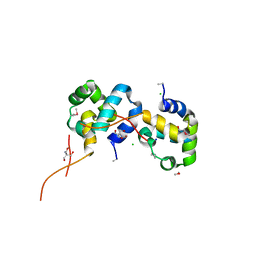 | | The Crystal Structure of the N-terminal domain of a RpiR Transcriptional Regulator from Staphylococcus epidermidis to 1.4A | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-METHOXYETHANOL, CHLORIDE ION, ... | | Authors: | Stein, A.J, Sather, A, Borovilos, M, Bargassa, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-09-02 | | Release date: | 2009-09-15 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Crystal Structure of the N-terminal domain of a RpiR Transcriptional Regulator from Staphylococcus epidermidis to 1.4A
To be Published
|
|
4R0T
 
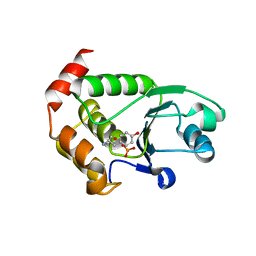 | | Crystal structure of P. aeruginosa TpbA (C132S) in complex with pTyr | | Descriptor: | PHOSPHATE ION, Protein tyrosine phosphatase TpbA, TYROSINE | | Authors: | Xu, K, Li, S, Wang, Y, Bartlam, M. | | Deposit date: | 2014-08-01 | | Release date: | 2015-05-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.603 Å) | | Cite: | Structural and Biochemical Analysis of Tyrosine Phosphatase Related to Biofilm Formation A (TpbA) from the Opportunistic Pathogen Pseudomonas aeruginosa PAO1
Plos One, 10
|
|
2XF6
 
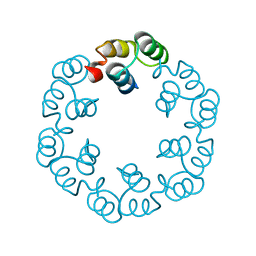 | | Crystal structure of Bacillus subtilis SPP1 phage gp23.1, a putative chaperone. | | Descriptor: | GP23.1 | | Authors: | Veesler, D, Blangy, S, Lichiere, J, Ortiz-Lombardia, M, Tavares, P, Campanacci, V, Cambillau, C. | | Deposit date: | 2010-05-20 | | Release date: | 2010-08-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Crystal Structure of Bacillus Subtilis Spp1 Phage Gp23.1, A Putative Chaperone.
Protein Sci., 19, 2010
|
|
4R0S
 
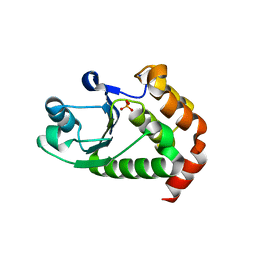 | | Crystal structure of P. aeruginosa TpbA | | Descriptor: | GLYCEROL, PHOSPHATE ION, Protein tyrosine phosphatase TpbA | | Authors: | Xu, K, Li, S, Wang, Y, Bartlam, M. | | Deposit date: | 2014-08-01 | | Release date: | 2015-05-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural and Biochemical Analysis of Tyrosine Phosphatase Related to Biofilm Formation A (TpbA) from the Opportunistic Pathogen Pseudomonas aeruginosa PAO1
PLoS ONE, 10, 2015
|
|
2HG7
 
 | | Solution NMR structure of Phage-like element PBSX protein xkdW, Northeast Structural Genomics Consortium Target SR355 | | Descriptor: | Phage-like element PBSX protein xkdW | | Authors: | Liu, G, Parish, D, Xu, D, Atreya, H, Sukumaran, D, Ho, C.K, Jiang, M, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M, Swapna, G.V, Acton, T.B, Rost, B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-06-26 | | Release date: | 2006-08-22 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of Phage-like element PBSX protein xkdW, Northeast Structural Genomics Consortium Target SR355
TO BE PUBLISHED
|
|
2HJ0
 
 | | Crystal Structure of the Putative Alfa Subunit of Citrate Lyase in Complex with Citrate from Streptococcus mutans, Northeast Structural Genomics Target SmR12 . | | Descriptor: | CITRIC ACID, Putative citrate lyase, alfa subunit | | Authors: | Forouhar, F, Hussain, M, Jayaraman, S, Shastry, R, Janjua, H, Cunningham, K, Ma, L.C, Xiao, R, Liu, J, Baran, M, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-06-29 | | Release date: | 2006-08-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of the Putative Alfa Subunit of Citrate Lyase in Complex with Citrate from Streptococcus mutans, Northeast Structural Genomics Target SmR12 (CASP Target).
To be Published
|
|
2XYK
 
 | | Group II 2-on-2 Hemoglobin from the Plant Pathogen Agrobacterium tumefaciens | | Descriptor: | 2-ON-2 HEMOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Pesce, A, Nardini, M, LaBarre, M, Richard, C, Wittenberg, J.B, Wittenberg, B.A, Guertin, M, Bolognesi, M. | | Deposit date: | 2010-11-18 | | Release date: | 2010-12-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Characterization of a Group II 2/2 Hemoglobin from the Plant Pathogen Agrobacterium Tumefaciens.
Biochim.Biophys.Acta, 1814, 2011
|
|
2HRX
 
 | | X-Ray Crystal Structure of Protein DIP2367 from Corynebacterium diphtheriae. Northeast Structural Genomics Consortium Target CdR13. | | Descriptor: | Hypothetical protein | | Authors: | Kuzin, A.P, Su, M, Jayaraman, S, Ho, C.K, Janjua, H, Cunningham, K, Ma, L.C, Xiao, R, Liu, J, Baran, M, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-07-20 | | Release date: | 2006-08-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Three dimensional structure of conserved hypothetical protein from Corynebacterium diphtheriae at the resolution 1.9 A. Northeast Structural Genomics Consortium target CdR13.
TO BE PUBLISHED
|
|
3L1N
 
 | | Crystal structure of Mp1p ligand binding domain 2 complexd with palmitic acid | | Descriptor: | Cell wall antigen, PALMITIC ACID | | Authors: | Liao, S, Tung, E.T, Zheng, W, Chong, K, Xu, Y, Bartlam, M, Rao, Z, Yuen, K.Y. | | Deposit date: | 2009-12-14 | | Release date: | 2010-01-05 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of the Mp1p ligand binding domain 2 reveals its function as a fatty acid-binding protein.
J.Biol.Chem., 285, 2010
|
|
1WOF
 
 | | Crystal Structure Of SARS-CoV Mpro in Complex with an Inhibitor N1 | | Descriptor: | 3C-like proteinase, N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]-L-ALANYL-L-VALYL-N~1~-((1S)-4-ETHOXY-4-OXO-1-{[(3S)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE | | Authors: | Yang, H, Bartlam, M, Xue, X, Yang, K, Liang, W, Rao, Z. | | Deposit date: | 2004-08-18 | | Release date: | 2005-08-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design of Wide-Spectrum Inhibitors Targeting Coronavirus Main Proteases.
Plos Biol., 3, 2005
|
|
3LXF
 
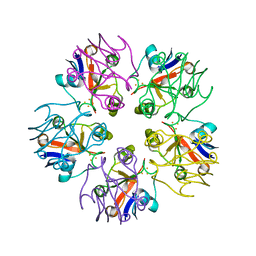 | | Crystal Structure of [2Fe-2S] Ferredoxin Arx from Novosphingobium aromaticivorans | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, Ferredoxin | | Authors: | Yang, W, Bell, S.G, Wang, H, Bartlam, M, Wong, L.L, Rao, Z. | | Deposit date: | 2010-02-25 | | Release date: | 2010-06-23 | | Last modified: | 2014-02-12 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular characterization of a class I P450 electron transfer system from Novosphingobium aromaticivorans DSM12444
J.Biol.Chem., 285, 2010
|
|
3LD1
 
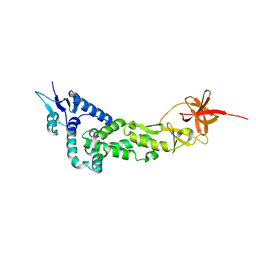 | | Crystal Structure of IBV Nsp2a | | Descriptor: | Replicase polyprotein 1a | | Authors: | Xu, Y, Cong, L, Wei, L, Fu, J, Chen, C, Yang, A, Tang, H, Bartlam, M, Rao, Z. | | Deposit date: | 2010-01-12 | | Release date: | 2011-05-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.498 Å) | | Cite: | IBV nsp2 is an endosome-associated protein and viral pathogenicity factor
To be Published
|
|
3LXI
 
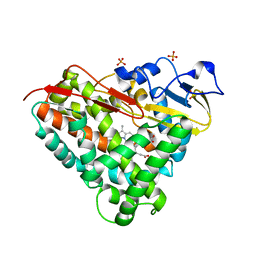 | | Crystal Structure of Camphor-Bound CYP101D1 | | Descriptor: | CAMPHOR, Cytochrome P450, PHOSPHATE ION, ... | | Authors: | Yang, W, Bell, S.G, Wang, H, Bartlam, M, Wong, L.L, Rao, Z. | | Deposit date: | 2010-02-25 | | Release date: | 2010-06-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular characterization of a class I P450 electron transfer system from Novosphingobium aromaticivorans DSM12444
J.Biol.Chem., 285, 2010
|
|
3LAE
 
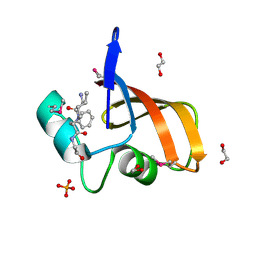 | | The crystal structure of a functionally unknown conserved protein from Haemophilus influenzae Rd KW20 | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHATE ION, UPF0053 protein HI0107, ... | | Authors: | Tan, K, Li, H, Bargassa, M, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-01-06 | | Release date: | 2010-01-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.453 Å) | | Cite: | The crystal structure of a functionally unknown conserved protein from Haemophilus influenzae Rd KW20
To be Published
|
|
3LXH
 
 | | Crystal Structure of Cytochrome P450 CYP101D1 | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Cytochrome P450, PHOSPHATE ION, ... | | Authors: | Yang, W, Bell, S.G, Wang, H, Bartlam, M, Wong, L.L, Rao, Z. | | Deposit date: | 2010-02-25 | | Release date: | 2010-06-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular characterization of a class I P450 electron transfer system from Novosphingobium aromaticivorans DSM12444
J.Biol.Chem., 285, 2010
|
|
3LXD
 
 | | Crystal Structure of Ferredoxin Reductase ArR from Novosphingobium aromaticivorans | | Descriptor: | FAD-dependent pyridine nucleotide-disulphide oxidoreductase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Yang, W, Bell, S.G, Wang, H, Bartlam, M, Wong, L.L, Rao, Z. | | Deposit date: | 2010-02-25 | | Release date: | 2010-06-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular characterization of a class I P450 electron transfer system from Novosphingobium aromaticivorans DSM12444
J.Biol.Chem., 285, 2010
|
|
3JU5
 
 | | Crystal Structure of Dimeric Arginine Kinase at 1.75-A Resolution | | Descriptor: | Arginine kinase, MAGNESIUM ION | | Authors: | Wu, X, Ye, S, Guo, S, Yan, W, Bartlam, M, Rao, Z. | | Deposit date: | 2009-09-14 | | Release date: | 2009-09-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for a reciprocating mechanism of negative cooperativity in dimeric phosphagen kinase activity
Faseb J., 24, 2010
|
|
3JU6
 
 | | Crystal Structure of Dimeric Arginine Kinase in Complex with AMPPNP and Arginine | | Descriptor: | ARGININE, Arginine kinase, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Wu, X, Ye, S, Guo, S, Yan, W, Bartlam, M, Rao, Z. | | Deposit date: | 2009-09-14 | | Release date: | 2009-09-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural basis for a reciprocating mechanism of negative cooperativity in dimeric phosphagen kinase activity
Faseb J., 24, 2010
|
|
