8K8K
 
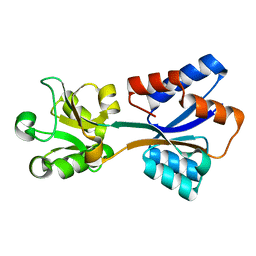 | | Structure of Klebsiella pneumonia ModA | | Descriptor: | Molybdate transporter periplasmic protein | | Authors: | Zhao, Q, Bartlam, M. | | Deposit date: | 2023-07-31 | | Release date: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural analysis of molybdate binding protein ModA from Klebsiella pneumoniae.
Biochem.Biophys.Res.Commun., 681, 2023
|
|
8K8L
 
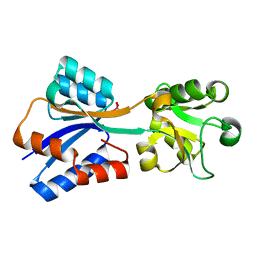 | |
8KHP
 
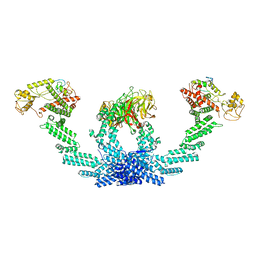 | | CULLIN3-KLHL22-RBX1 E3 ligase | | Descriptor: | Cullin-3, E3 ubiquitin-protein ligase RBX1, Kelch-like protein 22 | | Authors: | Su, M.-Y, Su, M.-Y. | | Deposit date: | 2023-08-22 | | Release date: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Cryo-EM structure of the KLHL22 E3 ligase bound to an oligomeric metabolic enzyme.
Structure, 31, 2023
|
|
8JKZ
 
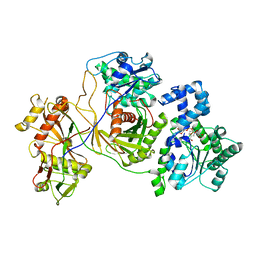 | |
8JL0
 
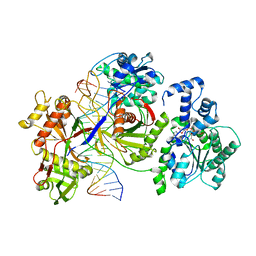 | | Cryo-EM structure of the prokaryotic SPARSA system complex | | Descriptor: | DNA (5'-D(P*AP*CP*GP*AP*CP*GP*TP*CP*TP*AP*AP*GP*AP*AP*AP*CP*CP*AP*TP*TP*AP*T)-3'), NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Piwi domain protein, ... | | Authors: | Xu, X, Zhen, X, Long, F. | | Deposit date: | 2023-06-02 | | Release date: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of antiphage immunity generated by a prokaryotic Argonaute-associated SPARSA system.
Nat Commun, 15, 2024
|
|
8VF6
 
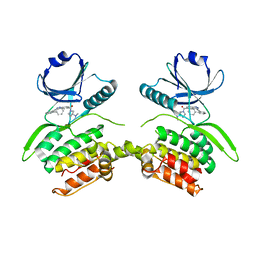 | | Crystal structure of Serine/threonine-protein kinase 33 (STK33) Kinase Domain in complex with inhibitor CDD-2211 | | Descriptor: | Serine/threonine-protein kinase 33, {3-[([1,1'-biphenyl]-2-yl)ethynyl]-1H-indazol-5-yl}[(3R)-3-(dimethylamino)pyrrolidin-1-yl]methanone | | Authors: | Ta, H.M, Kim, C, Ku, K.A, Matzuk, M.M. | | Deposit date: | 2023-12-21 | | Release date: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Reversible male contraception by targeted inhibition of serine/threonine kinase 33.
Science, 384, 2024
|
|
8KHR
 
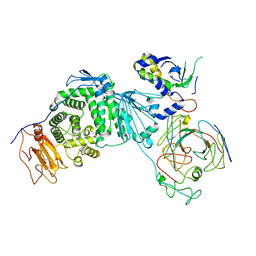 | | Cryo-EM structure of EBV gH/gL-gp42 in complex with fab 2C1 | | Descriptor: | 2C1 heavy chain, 2C1 light chain, Envelope glycoprotein H, ... | | Authors: | Fang, X.Y, Zhao, G.X, Zeng, M.S, Liu, Z. | | Deposit date: | 2023-08-22 | | Release date: | 2024-06-19 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Potent human monoclonal antibodies targeting Epstein-Barr virus gp42 reveal vulnerable sites for virus infection.
Cell Rep Med, 5, 2024
|
|
4OVF
 
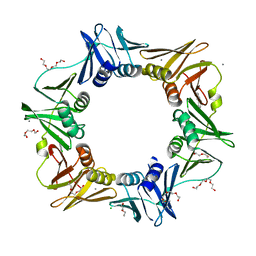 | | E. coli sliding clamp in complex with (R)-6-chloro-2,3,4,9-tetrahydro-1H-carbazole-2-carboxylic acid | | Descriptor: | (2R)-6-chloro-2,3,4,9-tetrahydro-1H-carbazole-2-carboxylic acid, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Yin, Z, Oakley, A.J. | | Deposit date: | 2014-02-21 | | Release date: | 2014-03-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Bacterial Sliding Clamp Inhibitors that Mimic the Sequential Binding Mechanism of Endogenous Linear Motifs.
J.Med.Chem., 58, 2015
|
|
4OVH
 
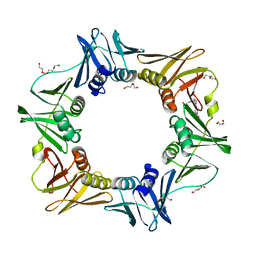 | | E. coli sliding clamp in complex with (R)-6-bromo-9-(2-(carboxymethylamino)-2-oxoethyl)-2,3,4,9-tetrahydro-1H-carbazole-2-carboxylic acid | | Descriptor: | (2R)-6-bromo-9-{2-[(carboxymethyl)amino]-2-oxoethyl}-2,3,4,9-tetrahydro-1H-carbazole-2-carboxylic acid, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Yin, Z, Oakley, A.J. | | Deposit date: | 2014-02-21 | | Release date: | 2014-03-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Bacterial Sliding Clamp Inhibitors that Mimic the Sequential Binding Mechanism of Endogenous Linear Motifs.
J.Med.Chem., 58, 2015
|
|
4IWX
 
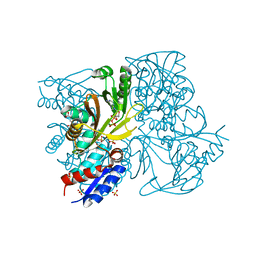 | | Rimk structure at 2.85A | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLUTAMIC ACID, Ribosomal protein S6 modification protein, ... | | Authors: | Shi, D, Zhao, G, Jin, Z, Allewell, N.M, Tuchman, M. | | Deposit date: | 2013-01-24 | | Release date: | 2013-05-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.854 Å) | | Cite: | Structure and function of Escherichia coli RimK, an ATP-grasp fold, l-glutamyl ligase enzyme.
Proteins, 81, 2013
|
|
4J1G
 
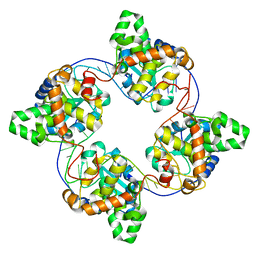 | | Leanyer orthobunyavirus nucleoprotein-ssRNA complex | | Descriptor: | Nucleocapsid, RNA (45-MER) | | Authors: | Niu, F, Shaw, N, Wang, Y, Jiao, L, Ding, W, Li, X, Zhu, P, Upur, H, Ouyang, S, Cheng, G, Liu, Z.J. | | Deposit date: | 2013-02-01 | | Release date: | 2013-04-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.789 Å) | | Cite: | Structure of the Leanyer orthobunyavirus nucleoprotein-RNA complex reveals unique architecture for RNA encapsidation.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
8OOJ
 
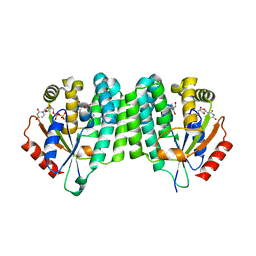 | | Crystal structure of dCK C4S-S74E mutant in complex with EdC and UDP | | Descriptor: | 4-azanyl-1-[(2~{R},4~{S},5~{R})-5-ethynyl-5-(hydroxymethyl)-4-oxidanyl-oxolan-2-yl]pyrimidin-2-one, Deoxycytidine kinase, URIDINE-5'-DIPHOSPHATE | | Authors: | Saez-Ayala, M, Rebuffet, E, Betzi, S, Morelli, X. | | Deposit date: | 2023-04-05 | | Release date: | 2023-12-20 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 4'-Ethynyl-2'-Deoxycytidine (EdC) Preferentially Targets Lymphoma and Leukemia Subtypes by Inducing Replicative Stress.
Mol.Cancer Ther., 23, 2024
|
|
7CPV
 
 | | Cryo-EM structure of 80S ribosome from mouse testis | | Descriptor: | 40S ribosomal protein S10, 40S ribosomal protein S11, 40S ribosomal protein S13, ... | | Authors: | Huo, Y.G, He, X, Jiang, T, Qin, Y, Guo, X.J, Sha, J.H. | | Deposit date: | 2020-08-08 | | Release date: | 2022-02-02 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | A male germ-cell-specific ribosome controls male fertility.
Nature, 612, 2022
|
|
7CPU
 
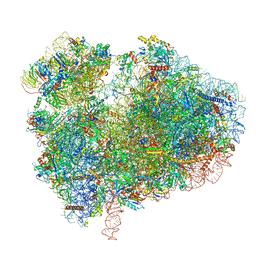 | | Cryo-EM structure of 80S ribosome from mouse kidney | | Descriptor: | 40S ribosomal protein S10, 40S ribosomal protein S11, 40S ribosomal protein S13, ... | | Authors: | Huo, Y.G, He, X, Jiang, T, Qin, Y, Guo, X.J, Sha, J.H. | | Deposit date: | 2020-08-08 | | Release date: | 2022-02-02 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | A male germ-cell-specific ribosome controls male fertility.
Nature, 612, 2022
|
|
8X83
 
 | |
8X82
 
 | |
8X84
 
 | |
8I5I
 
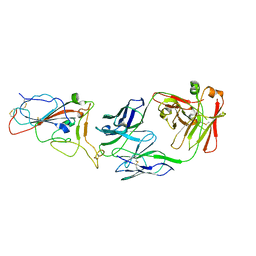 | |
8I5H
 
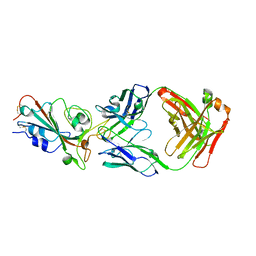 | |
5TWL
 
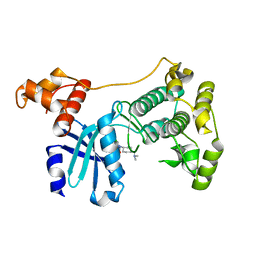 | | Structure of Maternal Embryonic Leucine Zipper Kinase | | Descriptor: | 9-(3,5-dichloro-4-hydroxyphenyl)-1-{trans-4-[(dimethylamino)methyl]cyclohexyl}-3,4-dihydropyrimido[5,4-c]quinolin-2(1H)-one, Maternal embryonic leucine zipper kinase | | Authors: | Seo, H.-S, Huang, H.-T, Gray, N.S, Dhe-Paganon, S. | | Deposit date: | 2016-11-14 | | Release date: | 2017-11-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | MELK is not necessary for the proliferation of basal-like breast cancer cells.
Elife, 6, 2017
|
|
5TWZ
 
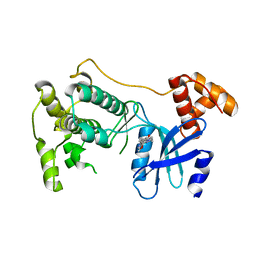 | | Structure of Maternal Embryonic Leucine Zipper Kinase | | Descriptor: | 7-{[2-methoxy-4-(1H-pyrazol-4-yl)benzoyl]amino}-2,3,4,5-tetrahydro-1H-3-benzazepinium, Maternal embryonic leucine zipper kinase | | Authors: | Seo, H.-Y, Dhe-Paganon, S. | | Deposit date: | 2016-11-15 | | Release date: | 2017-11-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.631 Å) | | Cite: | MELK is not necessary for the proliferation of basal-like breast cancer cells.
Elife, 6, 2017
|
|
7DNO
 
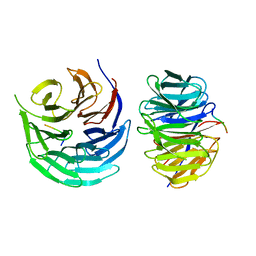 | | Characterization of Peptide Ligands Against WDR5 Isolated Using Phage Display Technique | | Descriptor: | CYS-ARG-THR-LEU-PRO-PHE, WD repeat-containing protein 5 | | Authors: | Cao, J, Cao, D, Xiong, B, Li, Y, Fan, T. | | Deposit date: | 2020-12-10 | | Release date: | 2021-02-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Phage-Display Based Discovery and Characterization of Peptide Ligands against WDR5.
Molecules, 26, 2021
|
|
7DHP
 
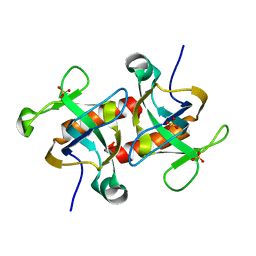 | | Crystal structure of MazF from Deinococcus radiodurans | | Descriptor: | Endoribonuclease MazF, SULFATE ION | | Authors: | Zhao, Y, Dai, J. | | Deposit date: | 2020-11-17 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | MazEF Toxin-Antitoxin System-Mediated DNA Damage Stress Response in Deinococcus radiodurans.
Front Genet, 12, 2021
|
|
7D0E
 
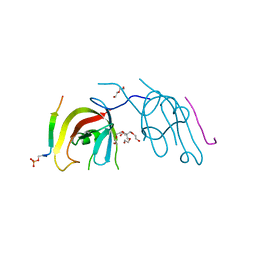 | | Crystal structure of FIP200 Claw/p-CCPG1 FIR2 | | Descriptor: | 3-(2-hydroxyethyloxy)-2-[2-(2-hydroxyethyloxy)ethoxymethyl]-2-(2-hydroxyethyloxymethyl)propan-1-ol, Cell cycle progression protein 1 FIR2, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Zhou, Z.X, Pan, L.F. | | Deposit date: | 2020-09-09 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Phosphorylation regulates the binding of autophagy receptors to FIP200 Claw domain for selective autophagy initiation.
Nat Commun, 12, 2021
|
|
7CZM
 
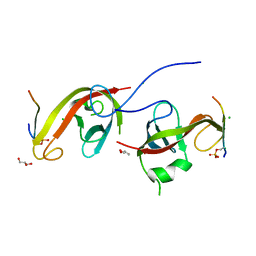 | | Crystal structure of FIP200 Claw/p-OPtineurin LIR complex | | Descriptor: | CHLORIDE ION, GLYCEROL, Optineurin LIR, ... | | Authors: | Zhou, Z.X, Pan, L.F. | | Deposit date: | 2020-09-09 | | Release date: | 2021-03-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Phosphorylation regulates the binding of autophagy receptors to FIP200 Claw domain for selective autophagy initiation.
Nat Commun, 12, 2021
|
|
