5YTE
 
 | | Large fragment of DNA Polymerase I from Thermus aquaticus in a closed ternary complex with with natural dT:dATP base pair | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, DNA (5'-D(*AP*AP*AP*TP*GP*GP*CP*GP*CP*CP*GP*TP*GP*GP*TP*C)-3'), DNA (5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*(DOC))-3'), ... | | Authors: | Zeng, H, Mondal, M, Song, R.Y, Zhang, J, Xia, B, Gao, Y.Q, Yi, C.Q. | | Deposit date: | 2017-11-17 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Unnatural Cytosine Bases Recognized as Thymines by DNA Polymerases by the Formation of the Watson-Crick Geometry.
Angew. Chem. Int. Ed. Engl., 58, 2019
|
|
1R5G
 
 | | Crystal Structure of MetAP2 complexed with A311263 | | Descriptor: | (2S,3R)-3-AMINO-2-HYDROXY-5-(ETHYLSULFANYL)PENTANOYL-((S)-(-)-(1-NAPHTHYL)ETHYL)AMIDE, MANGANESE (II) ION, Methionine aminopeptidase 2 | | Authors: | Sheppard, G.S, Wang, J, Kawai, M, BaMaung, N.Y, Craig, R.A, Erickson, S.A, Lynch, L, Patel, J, Yang, F, Searle, X.B, Lou, P, Park, C, Kim, K.H, Henkin, J, Lesniewski, R. | | Deposit date: | 2003-10-10 | | Release date: | 2004-10-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 3-Amino-2-hydroxyamides and related compounds as inhibitors of methionine aminopeptidase-2.
Bioorg.Med.Chem.Lett., 14, 2004
|
|
2MUV
 
 | | NOE-based model of the influenza A virus M2 (19-49) bound to drug 11 | | Descriptor: | (3s,5s,7s)-N-[(5-bromothiophen-2-yl)methyl]tricyclo[3.3.1.1~3,7~]decan-1-aminium, Matrix protein 2 | | Authors: | Wu, Y, Wang, J, DeGrado, W. | | Deposit date: | 2014-09-18 | | Release date: | 2014-12-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Flipping in the Pore: Discovery of Dual Inhibitors That Bind in Different Orientations to the Wild-Type versus the Amantadine-Resistant S31N Mutant of the Influenza A Virus M2 Proton Channel.
J.Am.Chem.Soc., 136, 2014
|
|
7JW3
 
 | | Crystal structure of Aedes aegypti Nibbler NTD domain | | Descriptor: | Exonuclease mut-7 homolog | | Authors: | Xie, W, Sowemimo, I, Hayashi, R, Wang, J, Brennecke, J, Ameres, S.L, Patel, D.J. | | Deposit date: | 2020-08-24 | | Release date: | 2021-01-20 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structure-function analysis of microRNA 3'-end trimming by Nibbler.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
1VYT
 
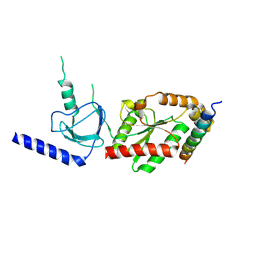 | | beta3 subunit complexed with aid | | Descriptor: | CALCIUM CHANNEL BETA-3 SUBUNIT, VOLTAGE-DEPENDENT L-TYPE CALCIUM CHANNEL ALPHA-1C SUBUNIT | | Authors: | Chen, Y.-H, Li, M.-H, Zhang, Y, He, L.-L, Yamada, Y, Fitzmaurice, A, Yang, S, Zhang, H, Tong, L, Yang, J. | | Deposit date: | 2004-05-07 | | Release date: | 2004-06-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis of the Alpha(1)-Beta Subunit Interaction of Voltage-Gated Ca(2+) Channels
Nature, 429, 2004
|
|
4NN2
 
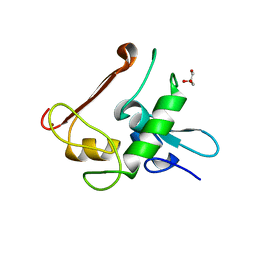 | | Protein Crystal Structure of Human Borjeson-Forssman-Lehmann Syndrome Associated Protein PHF6 | | Descriptor: | GLYCEROL, PHD finger protein 6, ZINC ION | | Authors: | Liu, Z, Li, F, Zhang, J, Mei, Y, Wu, J, Shi, Y. | | Deposit date: | 2013-11-16 | | Release date: | 2014-02-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.472 Å) | | Cite: | Crystal Structure of the second extended PHD domain of human PHF6 protein
J.Biol.Chem., 2014
|
|
1KUV
 
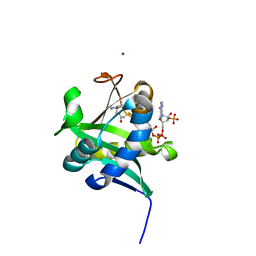 | | X-ray Crystallographic Studies of Serotonin N-acetyltransferase Catalysis and Inhibition | | Descriptor: | COA-S-ACETYL 5-BROMOTRYPTAMINE, MAGNESIUM ION, Serotonin N-acetyltransferase | | Authors: | Wolf, E, De Angelis, J, Khalil, E.M, Cole, P.A, Burley, S.K. | | Deposit date: | 2002-01-22 | | Release date: | 2002-03-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray crystallographic studies of serotonin N-acetyltransferase catalysis and inhibition.
J.Mol.Biol., 317, 2002
|
|
4CC6
 
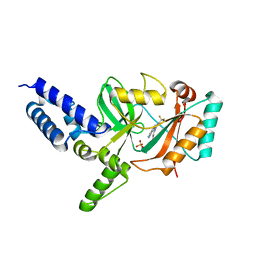 | | Fragment-Based Discovery of 6 Azaindazoles As Inhibitors of Bacterial DNA Ligase | | Descriptor: | 2-{[2-(1H-pyrazolo[3,4-c]pyridin-3-yl)-6-(trifluoromethyl)pyridin-4-yl]amino}ethanol, DNA LIGASE, SULFATE ION | | Authors: | Howard, S, Amin, N, Benowitz, A.B, Chiarparin, E, Cui, H, Deng, X, Heightman, T.D, Holmes, D.J, Hopkins, A, Huang, J, Jin, Q, Kreatsoulas, C, Martin, A.C.L, Massey, F, McCloskey, L, Mortenson, P.N, Pathuri, P, Tisi, D, Williams, P.A. | | Deposit date: | 2013-10-18 | | Release date: | 2014-06-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Fragment-Based Discovery of 6-Azaindazoles as Inhibitors of Bacterial DNA Ligase.
Acs Med.Chem.Lett., 4, 2013
|
|
6II0
 
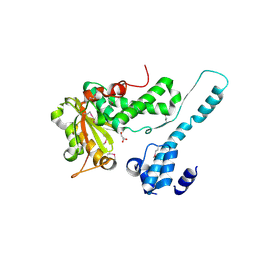 | | Crystal structure of the Makes Caterpillars Floppy (MCF)-Like effector of Vibrio vulnificus MO6-24/O | | Descriptor: | GLYCEROL, Putative RTX-toxin | | Authors: | Lee, Y, Kim, B.S, Choi, S, Lee, E.Y, Park, S, Hwang, J, Kwon, Y, Hyun, J, Lee, C, Eom, S.H, Kim, M.H. | | Deposit date: | 2018-10-03 | | Release date: | 2019-08-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Makes caterpillars floppy-like effector-containing MARTX toxins require host ADP-ribosylation factor (ARF) proteins for systemic pathogenicity.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
7JW2
 
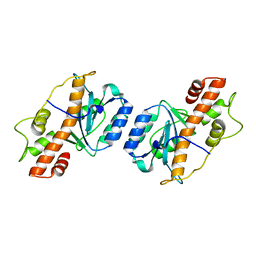 | | Crystal structure of Aedes aegypti Nibbler EXO domain | | Descriptor: | Exonuclease mut-7 homolog | | Authors: | Xie, W, Sowemimo, I, Hayashi, R, Wang, J, Brennecke, J, Ameres, S.L, Patel, D.J. | | Deposit date: | 2020-08-24 | | Release date: | 2021-01-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-function analysis of microRNA 3'-end trimming by Nibbler.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4CBY
 
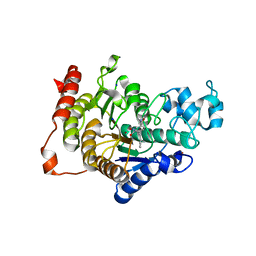 | | Design, synthesis, and biological evaluation of potent and selective Class IIa HDAC inhibitors as a potential therapy for Huntington's disease | | Descriptor: | (1R,2R,3R)-2-[4-(1,3-oxazol-5-yl)phenyl]-N-oxidanyl-3-phenyl-cyclopropane-1-carboxamide, HISTONE DEACETYLASE 4, SODIUM ION, ... | | Authors: | Burli, R.W, Luckhurst, C.A, Aziz, O, Matthews, K.L, Yates, D, Lyons, K.A, Beconi, M, McAllister, G, Breccia, P, Stott, A.J, Penrose, S.D, Wall, M, Lamers, M, Leonard, P, Mueller, I, Richardson, C.M, Jarvis, R, Stones, L, Hughes, S, Wishart, G, Haughan, A.F, O'Connell, C, Mead, T, McNeil, H, Vann, J, Mangette, J, Maillard, M, Beaumont, V, Munoz-Sanjuan, I, Dominguez, C. | | Deposit date: | 2013-10-17 | | Release date: | 2013-12-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Design, synthesis, and biological evaluation of potent and selective class IIa histone deacetylase (HDAC) inhibitors as a potential therapy for Huntington's disease.
J. Med. Chem., 56, 2013
|
|
2MZX
 
 | | CCR5-ECL2 helical structure, residues Q186-T195 | | Descriptor: | C-C chemokine receptor type 5 | | Authors: | Abayev, M, Anglister, J. | | Deposit date: | 2015-02-26 | | Release date: | 2015-04-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | An extended CCR5 ECL2 peptide forms a helix that binds HIV-1 gp120 through non-specific hydrophobic interactions.
Febs J., 282, 2015
|
|
5YTD
 
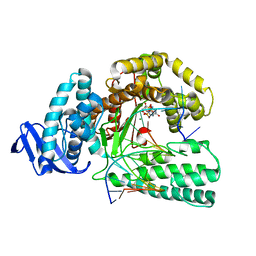 | | large fragment of DNA Polymerase I from Thermus aquaticus in a closed ternary complex with the natural base pair 5fC:dGTP | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA (5'-D(*AP*AP*AP*(5FC)P*GP*GP*CP*GP*CP*CP*GP*TP*GP*GP*TP*C)-3'), DNA (5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*(DOC))-3'), ... | | Authors: | Zeng, H, Mondal, M, Song, R.Y, Zhang, J, Xia, B, Gao, Y.Q, Yi, C.Q. | | Deposit date: | 2017-11-17 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Unnatural Cytosine Bases Recognized as Thymines by DNA Polymerases by the Formation of the Watson-Crick Geometry.
Angew. Chem. Int. Ed. Engl., 58, 2019
|
|
5YTC
 
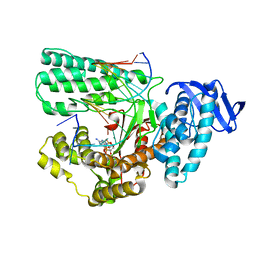 | | Large fragment of DNA Polymerase I from Thermus aquaticus in a closed ternary complex with the unnatural base M-fC pair with dATP in the active site | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, DNA (5'-D(*AP*AP*AP*(92F)P*GP*GP*CP*GP*CP*CP*GP*TP*GP*GP*TP*C)-3'), DNA (5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*(DOC)P*(DOC))-3'), ... | | Authors: | Zeng, H, Mondal, M, Song, R.Y, Zhang, J, Xia, B, Yi, C.Q. | | Deposit date: | 2017-11-17 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Unnatural Cytosine Bases Recognized as Thymines by DNA Polymerases by the Formation of the Watson-Crick Geometry.
Angew. Chem. Int. Ed. Engl., 58, 2019
|
|
4N41
 
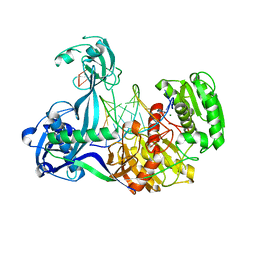 | | Structure of Thermus thermophilus Argonaute bound to guide DNA and 15-mer target DNA | | Descriptor: | 5'-D(*AP*AP*CP*CP*TP*AP*CP*TP*GP*CP*CP*TP*CP*G)-3', 5'-D(P*AP*CP*CP*TP*AP*CP*TP*AP*CP*CP*TP*CP*G)-3', 5'-D(P*TP*GP*AP*GP*GP*TP*AP*GP*TP*AP*GP*GP*TP*T*GP*TP*AP*TP*AP*GP*T)-3', ... | | Authors: | Sheng, G, Zhao, H, Wang, J, Rao, Y, Wang, Y. | | Deposit date: | 2013-10-08 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.248 Å) | | Cite: | Structure-based cleavage mechanism of Thermus thermophilus Argonaute DNA guide strand-mediated DNA target cleavage.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3R93
 
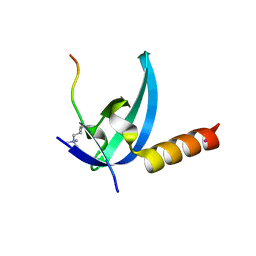 | | Crystal structure of the chromo domain of M-phase phosphoprotein 8 bound to H3K9Me3 peptide | | Descriptor: | H3K9Me3 peptide, M-phase phosphoprotein 8, UNKNOWN ATOM OR ION | | Authors: | Li, J, Li, Z, Ruan, J, Xu, C, Tong, Y, Pan, P.W, Tempel, W, Crombet, L, Min, J, Zang, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-03-24 | | Release date: | 2011-04-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.057 Å) | | Cite: | Structural basis for specific binding of human MPP8 chromodomain to histone H3 methylated at lysine 9.
Plos One, 6, 2011
|
|
5YTF
 
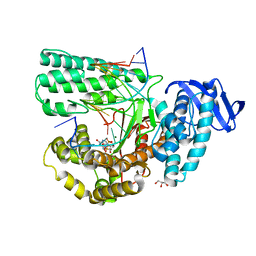 | | Structure of large fragment of DNA Polymerase I from Thermus aquaticus Host-Guest complex with the unnatural base M-fC pair with dA | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA (5'-D(*AP*AP*AP*CP*GP*GP*CP*GP*CP*CP*GP*(92F)P*GP*GP*TP*C)-3'), DNA (5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*(DOC))-3'), ... | | Authors: | Zeng, H, Mondal, M, Song, R.Y, Zhang, J, Xia, B, Gao, Y.Q, Yi, C.Q. | | Deposit date: | 2017-11-17 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Unnatural Cytosine Bases Recognized as Thymines by DNA Polymerases by the Formation of the Watson-Crick Geometry.
Angew. Chem. Int. Ed. Engl., 58, 2019
|
|
3RT0
 
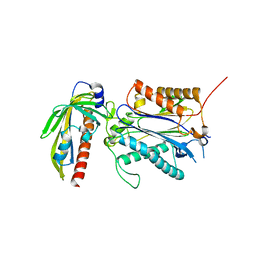 | | Crystal structure of PYL10-HAB1 complex in the absence of abscisic acid (ABA) | | Descriptor: | Abscisic acid receptor PYL10, MAGNESIUM ION, Protein phosphatase 2C 16 | | Authors: | Hao, Q, Yin, P, Li, W, Wang, L, Yan, C, Wang, J, Yan, N. | | Deposit date: | 2011-05-02 | | Release date: | 2011-06-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.113 Å) | | Cite: | The Molecular Basis of ABA-Independent Inhibition of PP2Cs by a Subclass of PYL Proteins
Mol.Cell, 42, 2011
|
|
1I27
 
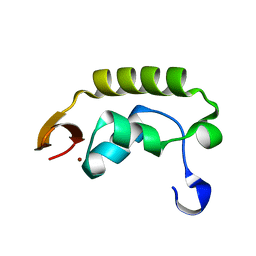 | | CRYSTAL STRUCTURE OF THE C-TERMINAL DOMAIN OF THE RAP74 SUBUNIT OF HUMAN TRANSCRIPTION FACTOR IIF (TFIIF) | | Descriptor: | TRANSCRIPTION FACTOR IIF, ZINC ION | | Authors: | Kamada, K, De Angelis, J, Roeder, R.G, Burley, S.K. | | Deposit date: | 2001-02-07 | | Release date: | 2001-03-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Crystal structure of the C-terminal domain of the RAP74 subunit of human transcription factor IIF.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
8P1R
 
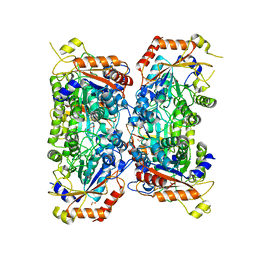 | | Bifidobacterium asteroides alpha-L-fucosidase (TT1819) catalytic mutant. | | Descriptor: | 1,2-ETHANEDIOL, Bifidobacterium asteroides alpha-L-fucosidase (TT1819) catalytic mutant, MAGNESIUM ION | | Authors: | Owen, C.D, Penner, M, Gascuena, A.N, Wu, H, Hernando, P.J, Monaco, S, Le Gall, G, Gardner, R, Ndeh, D, Urbanowicz, P.A, Spencer, D.I.R, Walsh, M.A, Angulo, J, Juge, N. | | Deposit date: | 2023-05-12 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.659 Å) | | Cite: | Exploring sequence, structure and function of microbial fucosidases from glycoside hydrolase GH29 family
To Be Published
|
|
5YTG
 
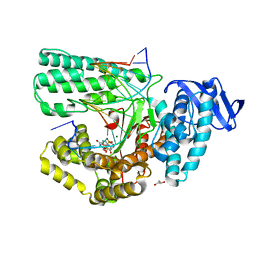 | | Structure of large fragment of DNA Polymerase I from Thermus aquaticus Host-Guest complex with the unnatural base I-fC pair with dA | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA (5'-D(*AP*AP*AP*CP*GP*GP*CP*GP*CP*CP*GP*(94O)P*GP*GP*TP*C)-3'), DNA (5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*(DOC))-3'), ... | | Authors: | Zeng, H, Mondal, M, Song, R.Y, Zhang, J, Xia, B, Gao, Y.Q, Yi, C.Q. | | Deposit date: | 2017-11-17 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Unnatural Cytosine Bases Recognized as Thymines by DNA Polymerases by the Formation of the Watson-Crick Geometry.
Angew. Chem. Int. Ed. Engl., 58, 2019
|
|
5YTH
 
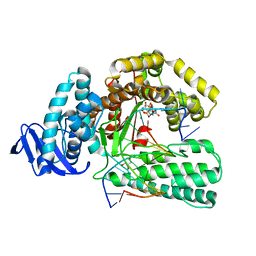 | | Structure of large fragment of DNA Polymerase I from Thermus aquaticus Host-Guest complex with the unnatural base M-fC pair with dG | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA (5'-D(*AP*AP*AP*CP*GP*GP*CP*GP*CP*CP*GP*(92F)P*GP*GP*TP*C)-3'), DNA (5'-D(*GP*AP*CP*CP*GP*CP*GP*GP*CP*GP*CP*(DOC))-3'), ... | | Authors: | Zeng, H, Mondal, M, Song, R.Y, Zhang, J, Xia, B, Gao, Y.Q, Yi, C.Q. | | Deposit date: | 2017-11-17 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Unnatural Cytosine Bases Recognized as Thymines by DNA Polymerases by the Formation of the Watson-Crick Geometry.
Angew. Chem. Int. Ed. Engl., 58, 2019
|
|
4AW2
 
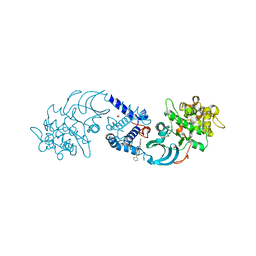 | | Crystal structure of CDC42 binding protein kinase alpha (MRCK alpha) | | Descriptor: | 1,2-ETHANEDIOL, 5,11-dimethyl-1-oxo-2,6-dihydro-1h-pyrido[4,3-b]carbazol-9-yl benzoate, SERINE/THREONINE-PROTEIN KINASE MRCK ALPHA | | Authors: | Elkins, J.M, Muniz, J.R.C, Tan, I, Leung, T, Lafanechere, L, Prudent, R, Abdul Azeez, K, Szklarz, M, Phillips, C, Wang, J, von Delft, F, Bountra, C, Edwards, A, Knapp, S. | | Deposit date: | 2012-05-30 | | Release date: | 2012-06-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Cdc42 Binding Protein Kinase Alpha (Mrck Alpha)
To be Published
|
|
3RT2
 
 | | Crystal structure of apo-PYL10 | | Descriptor: | Abscisic acid receptor PYL10 | | Authors: | Hao, Q, Yin, P, Li, W, Wang, L, Yan, C, Wang, J, Yan, N. | | Deposit date: | 2011-05-02 | | Release date: | 2011-06-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Molecular Basis of ABA-Independent Inhibition of PP2Cs by a Subclass of PYL Proteins
Mol.Cell, 42, 2011
|
|
3K6V
 
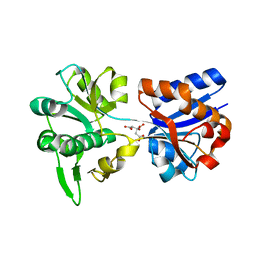 | | M. acetivorans Molybdate-Binding Protein (ModA) in Citrate-Bound Open Form | | Descriptor: | CITRIC ACID, Solute-binding protein MA_0280 | | Authors: | Chan, S, Giuroiu, I, Chernishof, I, Sawaya, M.R, Chiang, J, Gunsalus, R.P, Arbing, M.A, Perry, L.J. | | Deposit date: | 2009-10-09 | | Release date: | 2010-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Apo and ligand-bound structures of ModA from the archaeon Methanosarcina acetivorans
Acta Crystallogr.,Sect.F, 66, 2010
|
|
