8OQD
 
 | | Dirhodium tetraacetate/ribonuclease A adduct in the P3221 space group (1 h soaking) | | Descriptor: | CHLORIDE ION, FORMIC ACID, Ribonuclease pancreatic, ... | | Authors: | Loreto, D, Merlino, A, Maity, B, Ueno, T. | | Deposit date: | 2023-04-12 | | Release date: | 2023-06-14 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Cross-Linked Crystals of Dirhodium Tetraacetate/RNase A Adduct Can Be Used as Heterogeneous Catalysts.
Inorg.Chem., 62, 2023
|
|
8AJ4
 
 | | X-ray structure of lysozyme obtained upon reaction with [VIVO(malt)2] (Structure A') | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 8,8-bis($l^{1}-oxidanyl)-2,2'-dimethyl-8,8'-spirobi[3$l^{4},7,9-trioxa-8$l^{6}-vanadabicyclo[4.3.0]nona-1(6),2,4-triene], Lysozyme, ... | | Authors: | Paolillo, M, Merlino, A, Ferraro, G. | | Deposit date: | 2022-07-27 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Multiple and Variable Binding of Pharmacologically Active Bis(maltolato)oxidovanadium(IV) to Lysozyme.
Inorg.Chem., 61, 2022
|
|
8AJ3
 
 | | X-ray structure of lysozyme obtained upon reaction with [VIVO(malt)2] (Structure A) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 8,8,8,8-tetrakis($l^{1}-oxidanyl)-2-methyl-3,7,9-trioxa-8$l^{6}-vanadabicyclo[4.3.0]nona-1,5-diene, 8,8-bis($l^{1}-oxidanyl)-2,2'-dimethyl-8,8'-spirobi[3$l^{4},7,9-trioxa-8$l^{6}-vanadabicyclo[4.3.0]nona-1(6),2,4-triene], ... | | Authors: | Paolillo, M, Merlino, A, Ferraro, G. | | Deposit date: | 2022-07-27 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Multiple and Variable Binding of Pharmacologically Active Bis(maltolato)oxidovanadium(IV) to Lysozyme.
Inorg.Chem., 61, 2022
|
|
8AJ5
 
 | | X-ray structure of lysozyme obtained upon reaction with [VIVO(malt)2] (Structure B) | | Descriptor: | ACETATE ION, Lysozyme, NITRATE ION, ... | | Authors: | Paolillo, M, Merlino, A, Ferraro, G. | | Deposit date: | 2022-07-27 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Multiple and Variable Binding of Pharmacologically Active Bis(maltolato)oxidovanadium(IV) to Lysozyme.
Inorg.Chem., 61, 2022
|
|
8BRC
 
 | | Crystal structure of the adduct between human serum transferrin and cisplatin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, AMMONIA, FE (III) ION, ... | | Authors: | Troisi, R, Galardo, F, Ferraro, G, Sica, F, Merlino, A. | | Deposit date: | 2022-11-22 | | Release date: | 2023-01-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | Cisplatin Binding to Human Serum Transferrin: A Crystallographic Study.
Inorg.Chem., 62, 2023
|
|
5O7L
 
 | |
5O7S
 
 | | Crystal structure of a single chain monellin mutant (Y65R) pH 8.3 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Monellin chain B,Monellin chain A, SULFATE ION | | Authors: | Pica, A, Merlino, A. | | Deposit date: | 2017-06-09 | | Release date: | 2018-01-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | pH driven fibrillar aggregation of the super-sweet protein Y65R-MNEI: A step-by-step structural analysis.
Biochim. Biophys. Acta, 1862, 2017
|
|
5O7K
 
 | |
5O7R
 
 | |
5O7Q
 
 | | Crystal structure of a single chain monellin mutant (Y65R) pH 5.5 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Monellin chain B,Monellin chain A, SULFATE ION | | Authors: | Pica, A, Merlino, A. | | Deposit date: | 2017-06-09 | | Release date: | 2018-01-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | pH driven fibrillar aggregation of the super-sweet protein Y65R-MNEI: A step-by-step structural analysis.
Biochim. Biophys. Acta, 1862, 2017
|
|
5NJ1
 
 | |
5NA9
 
 | |
5NJ7
 
 | |
4NY5
 
 | |
4PPO
 
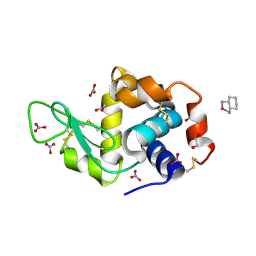 | | First Crystal Structure for an Oxaliplatin-Protein Complex | | Descriptor: | 1,2-ETHANEDIOL, CYCLOHEXANE-1(R),2(R)-DIAMINE-PLATINUM(II), Lysozyme C, ... | | Authors: | Messori, L, Merlino, A. | | Deposit date: | 2014-02-27 | | Release date: | 2015-01-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | The X-ray structure of the complex formed in the reaction between oxaliplatin and lysozyme.
Chem.Commun.(Camb.), 50, 2014
|
|
4OT4
 
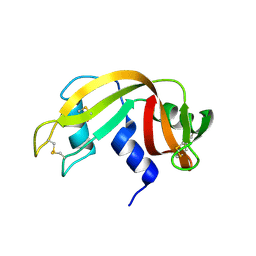 | |
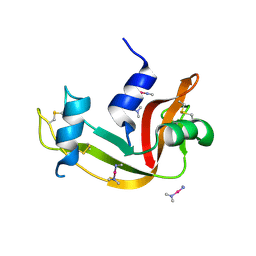
4QH3
 
 | |
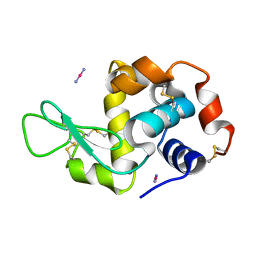
4QGZ
 
 | |
4QY9
 
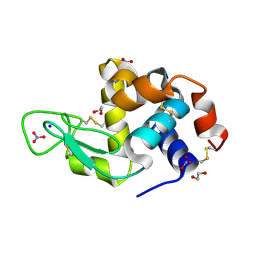 | | X-ray structure of the adduct between hen egg white lysozyme and Auoxo3, a cytotoxic gold(III) compound | | Descriptor: | 1,2-ETHANEDIOL, GOLD ION, Lysozyme C, ... | | Authors: | Russo Krauss, I, Merlino, A. | | Deposit date: | 2014-07-24 | | Release date: | 2014-11-05 | | Last modified: | 2014-11-19 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Interactions of gold-based drugs with proteins: the structure and stability of the adduct formed in the reaction between lysozyme and the cytotoxic gold(iii) compound Auoxo3.
Dalton Trans, 43, 2014
|
|
5OBE
 
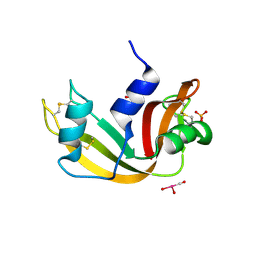 | | X-ray structure of the adduct formed upon reaction of ribonuclease A with the compound fac-[RuII(CO)3Cl2(N3-MBI), MBI=methyl-benzimidazole | | Descriptor: | PHOSPHATE ION, Ribonuclease pancreatic, pentakis(oxidaniumyl)-(oxidaniumylidynemethyl)ruthenium, ... | | Authors: | Pontillo, N, Ferraro, G, Merlino, A. | | Deposit date: | 2017-06-26 | | Release date: | 2017-07-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Ru-Based CO releasing molecules with azole ligands: interaction with proteins and the CO release mechanism disclosed by X-ray crystallography.
Dalton Trans, 46, 2017
|
|
5OLE
 
 | |
5OLD
 
 | |
5OB7
 
 | | X-ray structure of the adduct formed upon reaction of lysozyme with the compound fac-[RuII(CO)3Cl2(N3-IM), IM=imidazole (crystal 2) | | Descriptor: | CHLORIDE ION, GLYCEROL, Lysozyme C, ... | | Authors: | Pontillo, N, Ferraro, G, Merlino, A. | | Deposit date: | 2017-06-26 | | Release date: | 2017-07-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Ru-Based CO releasing molecules with azole ligands: interaction with proteins and the CO release mechanism disclosed by X-ray crystallography.
Dalton Trans, 46, 2017
|
|
5OBC
 
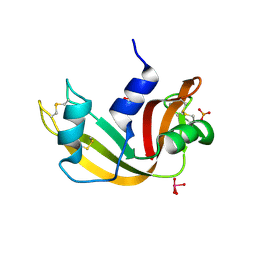 | | X-ray structure of the adduct formed upon reaction of ribonuclease A with the compound fac-[RuII(CO)3Cl2(N3-IM), IM=imidazole | | Descriptor: | PHOSPHATE ION, Ribonuclease pancreatic, pentakis(oxidaniumyl)-(oxidaniumylidynemethyl)ruthenium, ... | | Authors: | Pontillo, N, Ferraro, G, Merlino, A. | | Deposit date: | 2017-06-26 | | Release date: | 2017-07-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Ru-Based CO releasing molecules with azole ligands: interaction with proteins and the CO release mechanism disclosed by X-ray crystallography.
Dalton Trans, 46, 2017
|
|
5OB9
 
 | | X-ray structure of the adduct formed upon reaction of lysozyme with the compound fac-[RuII(CO)3Cl2(N3-MIM), MIM=methyl-imidazole (crystals grown using ethylene glycol | | Descriptor: | 1,2-ETHANEDIOL, Lysozyme C, NITRATE ION, ... | | Authors: | Pontillo, N, Ferraro, G, Merlino, A. | | Deposit date: | 2017-06-26 | | Release date: | 2017-07-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Ru-Based CO releasing molecules with azole ligands: interaction with proteins and the CO release mechanism disclosed by X-ray crystallography.
Dalton Trans, 46, 2017
|
|
